
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jiaying Lin | -- | 2759 | 2022-12-28 03:38:14 | | | |
| 2 | Vivi Li | Meta information modification | 2759 | 2022-12-29 01:53:42 | | |
Video Upload Options
Drug-induced liver injury (DILI) is a major cause of the withdrawal of pre-marketed drugs, typically attributed to oxidative stress, mitochondrial damage, disrupted bile acid homeostasis, and innate immune-related inflammation. DILI can be divided into intrinsic and idiosyncratic DILI with cholestatic liver injury as an important manifestation. The diagnosis of DILI remains a challenge today and relies on clinical judgment and knowledge of the insulting agent. Early prediction of hepatotoxicity is an important but still unfulfilled component of drug development. In response, in silico modeling has shown good potential to fill the missing puzzle. Computer algorithms, with machine learning and artificial intelligence as a representative, can be established to initiate a reaction on the given condition to predict DILI.
1. Introduction
2. The Prediction of DILI by In Silico Models
2.1. Knowledge-Based Prediction
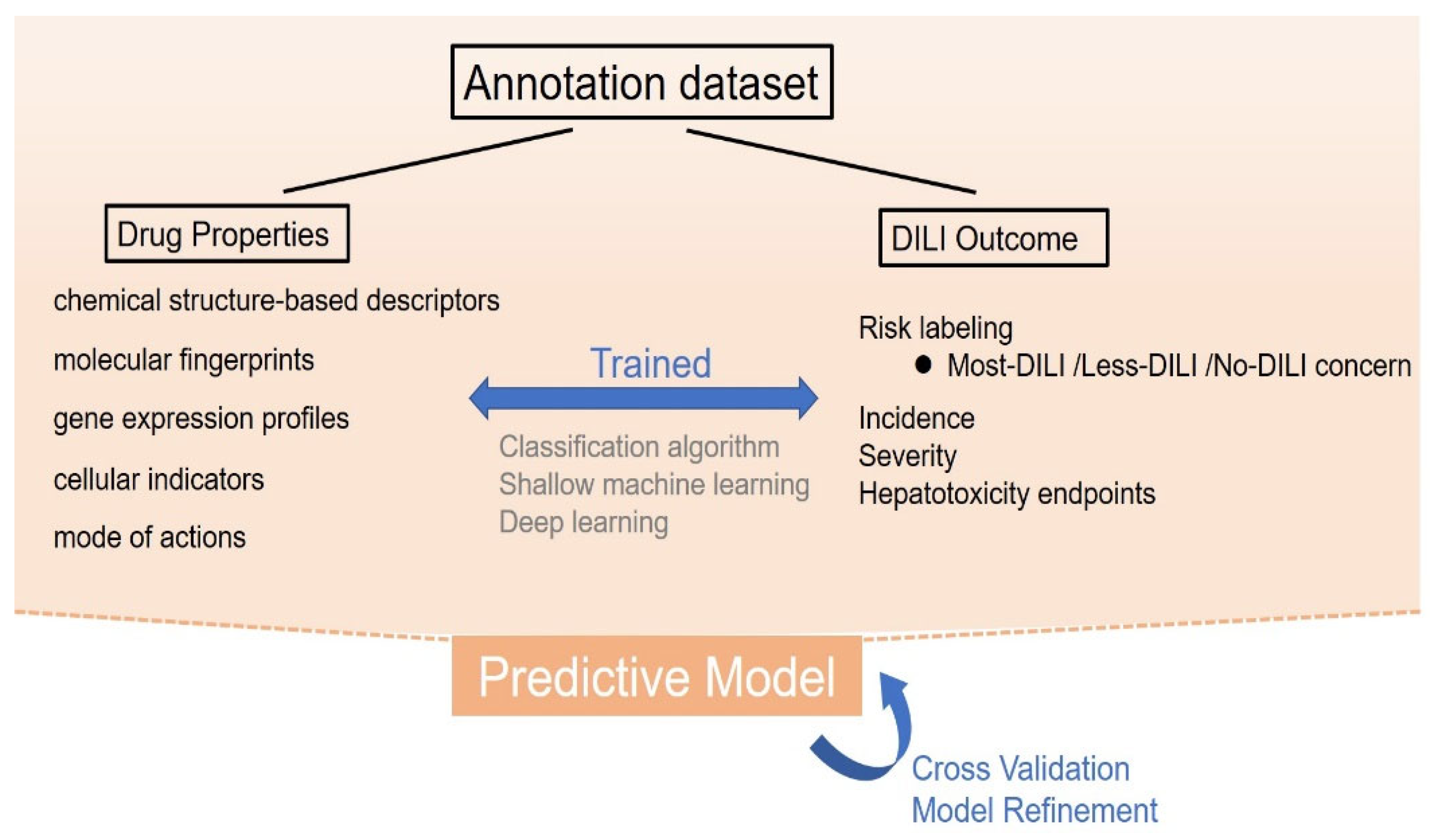
2.1.1. Cheminformatics-Based Model
Expert Knowledge Approaches
2.1.2. Bioactivity-Based Model
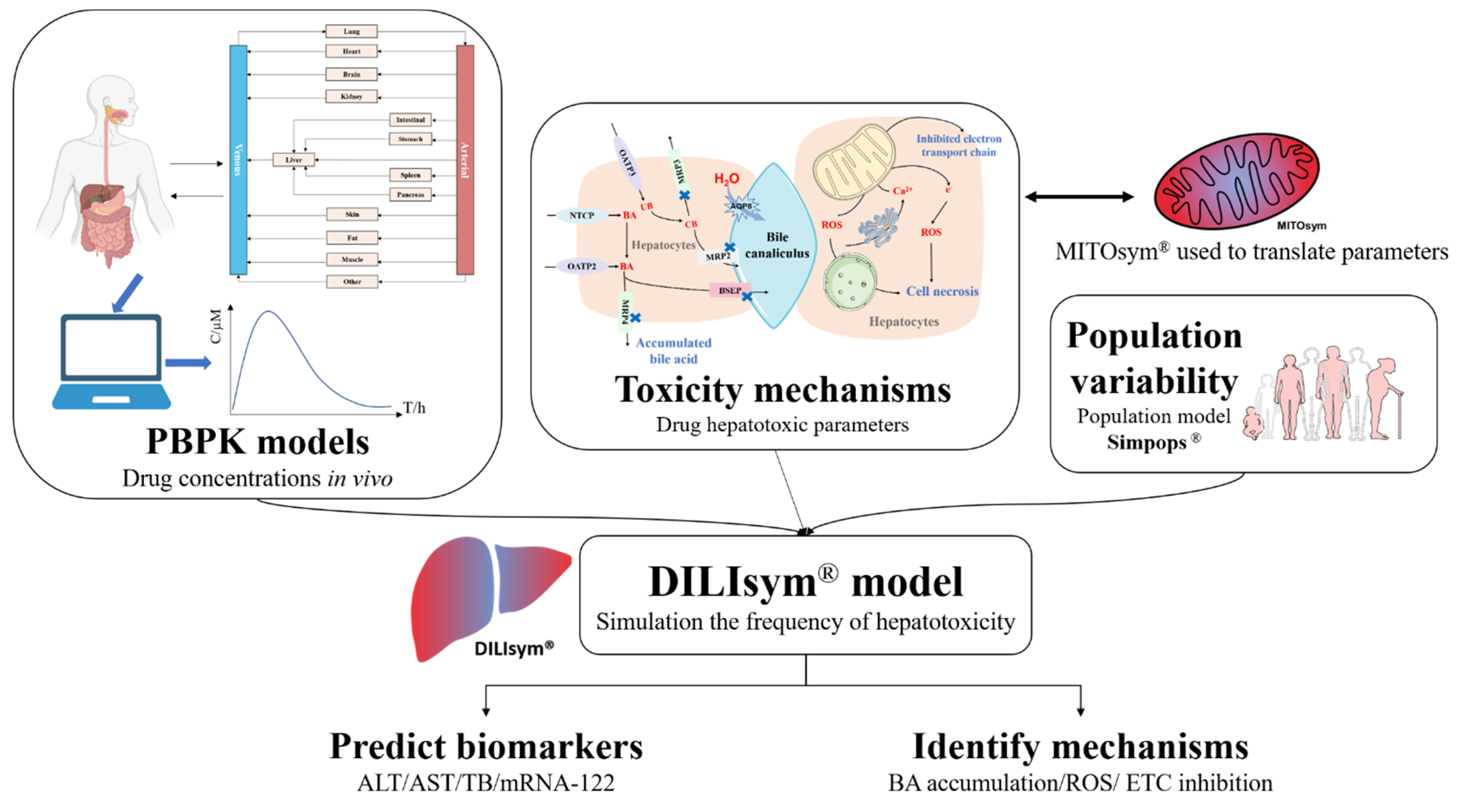
2.2. Mechanism-Based Prediction
References
- De Abajo, F.J.; Montero, D.; Madurga, M.; García Rodríguez, L.A. Acute and clinically relevant drug-induced liver injury: A population based case-control study. Br. J. Clin. Pharmacol. 2004, 58, 71–80.
- Shen, T.; Liu, Y.; Shang, J.; Xie, Q.; Li, J.; Yan, M.; Xu, J.; Niu, J.; Liu, J.; Watkins, P.B.; et al. Incidence and Etiology of Drug-Induced Liver Injury in Mainland China. Gastroenterology 2019, 156, 2230–2241.e2211.
- Bégaud, B.; Martin, K.; Haramburu, F.; Moore, N. Rates of spontaneous reporting of adverse drug reactions in France. JAMA 2002, 288, 1588.
- Björnsson, E.S. Epidemiology and risk factors for idiosyncratic drug-induced liver injury. Semin. Liver Dis. 2014, 34, 115–122.
- Sgro, C.; Clinard, F.; Ouazir, K.; Chanay, H.; Allard, C.; Guilleminet, C.; Lenoir, C.; Lemoine, A.; Hillon, P. Incidence of drug-induced hepatic injuries: A French population-based study. Hepatology 2002, 36, 451–455.
- Andrade, R.J.; Chalasani, N.; Björnsson, E.S.; Suzuki, A.; Kullak-Ublick, G.A.; Watkins, P.B.; Devarbhavi, H.; Merz, M.; Lucena, M.I.; Kaplowitz, N.; et al. Drug-induced liver injury. Nat. Rev. Dis. Prim. 2019, 5, 58.
- Garcia-Cortes, M.; Robles-Diaz, M.; Stephens, C.; Ortega-Alonso, A.; Lucena, M.I.; Andrade, R.J. Drug induced liver injury: An update. Arch. Toxicol. 2020, 94, 3381–3407.
- Donato, M.T.; Gallego-Ferrer, G.; Tolosa, L. In Vitro Models for Studying Chronic Drug-Induced Liver Injury. Int. J. Mol. Sci. 2022, 23, 11428.
- Segovia-Zafra, A.; Di Zeo-Sánchez, D.E.; López-Gómez, C.; Pérez-Valdés, Z.; García-Fuentes, E.; Andrade, R.J.; Lucena, M.I.; Villanueva-Paz, M. Preclinical models of idiosyncratic drug-induced liver injury (iDILI): Moving towards prediction. Acta Pharm. Sin. B 2021, 11, 3685–3726.
- Ozawa, S.; Miura, T.; Terashima, J.; Habano, W.; Ishida, S. Recent Progress in Prediction Systems for Drug-induced Liver Injury Using In vitro Cell Culture. Drug Metab. Lett. 2021, 14, 25–40.
- Kenna, J.G.; Uetrecht, J. Do In Vitro Assays Predict Drug Candidate Idiosyncratic Drug-Induced Liver Injury Risk? Drug Metab. Dispos. 2018, 46, 1658–1669.
- Fernández-Murga, M.L.; Petrov, P.D.; Conde, I.; Castell, J.V.; Goméz-Lechón, M.J.; Jover, R. Advances in drug-induced cholestasis: Clinical perspectives, potential mechanisms and in vitro systems. Food Chem. Toxicol. 2018, 120, 196–212.
- Gómez-Lechón, M.J.; Tolosa, L.; Donato, M.T. Metabolic activation and drug-induced liver injury: In vitro approaches for the safety risk assessment of new drugs. J. Appl. Toxicol. 2016, 36, 752–768.
- Hasegawa, M.; Kawai, K.; Mitsui, T.; Taniguchi, K.; Monnai, M.; Wakui, M.; Ito, M.; Suematsu, M.; Peltz, G.; Nakamura, M.; et al. The reconstituted ‘humanized liver’ in TK-NOG mice is mature and functional. Biochem. Biophys. Res. Commun. 2011, 405, 405–410.
- Azuma, H.; Paulk, N.; Ranade, A.; Dorrell, C.; Al-Dhalimy, M.; Ellis, E.; Strom, S.; Kay, M.A.; Finegold, M.; Grompe, M. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat. Biotechnol. 2007, 25, 903–910.
- Naritomi, Y.; Sanoh, S.; Ohta, S. Chimeric mice with humanized liver: Application in drug metabolism and pharmacokinetics studies for drug discovery. Drug Metab. Pharmacokinet. 2018, 33, 31–39.
- Bissig, K.D.; Han, W.; Barzi, M.; Kovalchuk, N.; Ding, L.; Fan, X.; Pankowicz, F.P.; Zhang, Q.Y.; Ding, X. P450-Humanized and Human Liver Chimeric Mouse Models for Studying Xenobiotic Metabolism and Toxicity. Drug Metab. Dispos. 2018, 46, 1734–1744.
- Fernandez-Checa, J.C.; Bagnaninchi, P.; Ye, H.; Sancho-Bru, P.; Falcon-Perez, J.M.; Royo, F.; Garcia-Ruiz, C.; Konu, O.; Miranda, J.; Lunov, O.; et al. Advanced preclinical models for evaluation of drug-induced liver injury—Consensus statement by the European Drug-Induced Liver Injury Network . J. Hepatol. 2021, 75, 935–959.
- Andrade, R.J.; Robles, M.; Lucena, M.I. Rechallenge in drug-induced liver injury: The attractive hazard. Expert Opin. Drug Saf. 2009, 8, 709–714.
- Fontana, R.J.; Watkins, P.B.; Bonkovsky, H.L.; Chalasani, N.; Davern, T.; Serrano, J.; Rochon, J. Drug-Induced Liver Injury Network (DILIN) prospective study: Rationale, design and conduct. Drug Saf. 2009, 32, 55–68.
- Buchanan, J.; Li, M.; Ni, X.; Wildfire, J. A New Paradigm for Safety Data Signal Detection and Evaluation Using Open-Source Software Created by an Interdisciplinary Working Group. Ther. Innov. Regul. Sci. 2021, 55, 1214–1219.
- Przybylak, K.R.; Cronin, M.T.D. In silico models for drug-induced liver injury--current status. Expert Opin. Drug Metab. Toxicol. 2012, 8, 201–217.
- Aguiar-Pulido, V.; Gestal, M.; Cruz-Monteagudo, M.; Rabuñal, J.R.; Dorado, J.; Munteanu, C.R. Evolutionary computation and QSAR research. Curr. Comput. Aided Drug Des. 2013, 9, 206–225.
- Idakwo, G.; Luttrell, J.; Chen, M.; Hong, H.; Zhou, Z.; Gong, P.; Zhang, C. A review on machine learning methods for in silico toxicity prediction. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2018, 36, 169–191.
- Vall, A.; Sabnis, Y.; Shi, J.; Class, R.; Hochreiter, S.; Klambauer, G. The Promise of AI for DILI Prediction. Front. Artif. Intell. 2021, 4, 638410.
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14.
- Limban, C.; Nuţă, D.C.; Chiriţă, C.; Negreș, S.; Arsene, A.L.; Goumenou, M.; Karakitsios, S.P.; Tsatsakis, A.M.; Sarigiannis, D.A. The use of structural alerts to avoid the toxicity of pharmaceuticals. Toxicol. Rep. 2018, 5, 943–953.
- Egan, W.J.; Zlokarnik, G.; Grootenhuis, P.D.J. In silico prediction of drug safety: Despite progress there is abundant room for improvement. Drug Discov. Today Technol. 2004, 1, 381–387.
- Marchant, C.A.; Briggs, K.A.; Long, A. In silico tools for sharing data and knowledge on toxicity and metabolism: Derek for windows, meteor, and vitic. Toxicol. Mech. Methods 2008, 18, 177–187.
- Greene, N.; Fisk, L.; Naven, R.T.; Note, R.R.; Patel, M.L.; Pelletier, D.J. Developing structure-activity relationships for the prediction of hepatotoxicity. Chem. Res. Toxicol. 2010, 23, 1215–1222.
- Zhu, X.; Kruhlak, N.L. Construction and analysis of a human hepatotoxicity database suitable for QSAR modeling using post-market safety data. Toxicology 2014, 321, 62–72.
- Lamb, J. The Connectivity Map: A new tool for biomedical research. Nat. Rev. Cancer 2007, 7, 54–60.
- Rueda-Zárate, H.A.; Imaz-Rosshandler, I.; Cárdenas-Ovando, R.A.; Castillo-Fernández, J.E.; Noguez-Monroy, J.; Rangel-Escareño, C. A computational toxicogenomics approach identifies a list of highly hepatotoxic compounds from a large microarray database. PLoS ONE 2017, 12, e0176284.
- Liu, X.; Zheng, D.; Zhong, Y.; Xia, Z.; Luo, H.; Weng, Z. Machine-Learning Prediction of Oral Drug-Induced Liver Injury (DILI) via Multiple Features and Endpoints. Biomed. Res. Int. 2020, 2020, 4795140.
- Li, T.; Tong, W.; Roberts, R.; Liu, Z.; Thakkar, S. Deep Learning on High-Throughput Transcriptomics to Predict Drug-Induced Liver Injury. Front. Bioeng. Biotechnol. 2020, 8, 562677.
- Zhu, X.-W.; Sedykh, A.; Liu, S.-S. Hybrid in silico models for drug-induced liver injury using chemical descriptors and in vitro cell-imaging information. J. Appl. Toxicol. 2014, 34, 281–288.
- Puri, M. Automated Machine Learning Diagnostic Support System as a Computational Biomarker for Detecting Drug-Induced Liver Injury Patterns in Whole Slide Liver Pathology Images. Assay Drug Dev. Technol. 2020, 18, 1–10.
- Wu, L.; Liu, Z.; Auerbach, S.; Huang, R.; Chen, M.; McEuen, K.; Xu, J.; Fang, H.; Tong, W. Integrating Drug’s Mode of Action into Quantitative Structure-Activity Relationships for Improved Prediction of Drug-Induced Liver Injury. J. Chem. Inf. Model. 2017, 57, 1000–1006.
- Watkins, P.B. The DILI-sim Initiative: Insights into Hepatotoxicity Mechanisms and Biomarker Interpretation. Clin. Transl. Sci. 2019, 12, 122–129.
- Yang, Y.; Nadanaciva, S.; Will, Y.; Woodhead, J.L.; Howell, B.A.; Watkins, P.B.; Siler, S.Q. MITOsym®: A Mechanistic, Mathematical Model of Hepatocellular Respiration and Bioenergetics. Pharm. Res. 2015, 32, 1975–1992.
- Watkins, P.B. Quantitative Systems Toxicology Approaches to Understand and Predict Drug-Induced Liver Injury. Clin. Liver Dis. 2020, 24, 49–60.
- Shoda, L.K.; Battista, C.; Siler, S.Q.; Pisetsky, D.S.; Watkins, P.B.; Howell, B.A. Mechanistic Modelling of Drug-Induced Liver Injury: Investigating the Role of Innate Immune Responses. Gene Regul. Syst. Biol. 2017, 11, 1177625017696074.
- Shoda, L.K.M.; Woodhead, J.L.; Siler, S.Q.; Watkins, P.B.; Howell, B.A. Linking physiology to toxicity using DILIsym®, a mechanistic mathematical model of drug-induced liver injury. Biopharm. Drug Dispos. 2014, 35, 33–49.
- Chung, J.-Y.; Longo, D.M.; Watkins, P.B. A Rapid Method to Estimate Hepatocyte Loss Due to Drug-Induced Liver Injury. Clin. Pharmacol. Ther. 2019, 105, 746–753.
- Teorell, T. Kinetics of distribution of substances administered to body. Arch. Int. Pharmacodyn. Ther. 1937, 57, 205–225.
- Kuepfer, L.; Niederalt, C.; Wendl, T.; Schlender, J.F.; Willmann, S.; Lippert, J.; Block, M.; Eissing, T.; Teutonico, D. Applied Concepts in PBPK Modeling: How to Build a PBPK/PD Model. CPT Pharmacomet. Syst. Pharmacol. 2016, 5, 516–531.
- Zhuang, X.; Lu, C. PBPK modeling and simulation in drug research and development. Acta Pharm. Sin. B 2016, 6, 430–440.
- Rowland, M.; Peck, C.; Tucker, G. Physiologically-based pharmacokinetics in drug development and regulatory science. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 45–73.
- Jin, Z.; He, Q.; Zhu, X.; Zhu, M.; Wang, Y.; Wu, X.A.; Lv, Q.; Xiang, X. Application of physiologically based pharmacokinetic modelling for the prediction of drug-drug interactions involving anlotinib as a perpetrator of cytochrome P450 enzymes. Basic Clin. Pharmacol. Toxicol. 2022, 130, 592–605.
- Woodhead, J.L.; Howell, B.A.; Yang, Y.; Harrill, A.H.; Clewell, H.J.; Andersen, M.E.; Siler, S.Q.; Watkins, P.B. An Analysis of N-Acetylcysteine Treatment for Acetaminophen Overdose Using a Systems Model of Drug-Induced Liver Injury. J. Pharmacol. Exp. Ther. 2012, 342, 529–540.
- Smith, B.; Rowe, J.; Watkins, P.B.; Ashina, M.; Woodhead, J.L.; Sistare, F.D.; Goadsby, P.J. Mechanistic Investigations Support Liver Safety of Ubrogepant. Toxicol. Sci. 2020, 177, 84–93.
- Longo, D.M.; Shoda, L.K.M.; Howell, B.A.; Coric, V.; Berman, R.M.; Qureshi, I.A. Assessing Effects of BHV-0223 40 mg Zydis Sublingual Formulation and Riluzole 50 mg Oral Tablet on Liver Function Test Parameters Utilizing DILIsym. Toxicol. Sci. 2020, 175, 292–300.
- Woodhead, J.L.; Yang, K.; Oldach, D.; MacLauchlin, C.; Fernandes, P.; Watkins, P.B.; Siler, S.Q.; Howell, B.A. Analyzing the Mechanisms Behind Macrolide Antibiotic-Induced Liver Injury Using Quantitative Systems Toxicology Modeling. Pharm. Res. 2019, 36, 48.
- Beaudoin, J.J.; Brock, W.J.; Watkins, P.B.; Brouwer, K.L.R. Quantitative Systems Toxicology Modeling Predicts that Reduced Biliary Efflux Contributes to Tolvaptan Hepatotoxicity. Clin. Pharmacol. Ther. 2021, 109, 433–442.




