Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Fan, W.; Yang, C.; Hou, X.; Wan, J.; Liao, B. Sinoatrial Node in Single-Cell RNA Sequencing. Encyclopedia. Available online: https://encyclopedia.pub/entry/38331 (accessed on 07 February 2026).
Fan W, Yang C, Hou X, Wan J, Liao B. Sinoatrial Node in Single-Cell RNA Sequencing. Encyclopedia. Available at: https://encyclopedia.pub/entry/38331. Accessed February 07, 2026.
Fan, Wei, Chao Yang, Xiaojie Hou, Juyi Wan, Bin Liao. "Sinoatrial Node in Single-Cell RNA Sequencing" Encyclopedia, https://encyclopedia.pub/entry/38331 (accessed February 07, 2026).
Fan, W., Yang, C., Hou, X., Wan, J., & Liao, B. (2022, December 08). Sinoatrial Node in Single-Cell RNA Sequencing. In Encyclopedia. https://encyclopedia.pub/entry/38331
Fan, Wei, et al. "Sinoatrial Node in Single-Cell RNA Sequencing." Encyclopedia. Web. 08 December, 2022.
Copy Citation
Normal cardiac automaticity is dependent on the pacemaker cells of the sinoatrial node (SAN). Insufficient cardiac pacemaking leads to the development of sick sinus syndrome (SSS). The development of more effective treatments for SSS, including biological pacemakers, requires further understanding of these genes and signaling pathways. Compared with genetic models and bulk RNA sequencing, single-cell RNA sequencing (scRNA-seq) technology promises to advance the understanding of cellular phenotype heterogeneity and molecular regulation during SAN development.
sinoatrial node
single-cell RNA sequencing
transcription factors
1. Introduction
The sinoatrial node (SAN) is a small population of cardiomyocytes that spontaneously fire to trigger each heartbeat. Abnormal SAN formation or function can lead to sick sinus syndrome (SSS), including sinus bradycardia, sinoatrial arrest/block, bradycardia–tachycardia syndrome, and sudden death [1]. Currently, no drugs for long-term use are effective for SSS treatment [2]. Development of more effective treatments for SSS, such as biological pacemakers, requires a comprehensive understanding of the genes and signaling pathways involved in the regulation of SAN development and function. Since the discovery of the SAN more than a century ago, studies have uncovered its intricate molecular structure and unique ion channels expressed within its myocytes. However, the molecular and cellular features that influence the pivotal functions of the SAN in the heart have not been fully elucidated [2].
The SAN is a subtle biological system that comprises multiple subdomains and cell types with distinct functions, including pacemaker cells (PCs) and several non-PCs [3]. Some previous studies that used genetically deficient mice (TBX3, TBX5, TBX18, SHOX2, NKX2-5, and PITX2) provided initial insights into SAN development and function [4][5][6][7][8]. Unfortunately, these studies are often limited by the small size and heterogeneous organization of the SAN, providing insufficient knowledge on gene regulation of the SAN [9]. Bulk RNA sequencing (RNA-seq) provides transcriptome data for the study of the SAN, further advancing SAN research. However, given the lack of specific molecular markers and the small size of the SAN, transcriptome data are often distorted and affected by other cells [10]. Despite significant progress in SAN research, the understanding of its cellular features, differentiation trajectories, and regulatory mechanisms is incomplete. Some obstacles in investigating the molecular features of the SAN include (1) the low total number of PCs in the heart, (2) the complex three-dimensional anatomy of the SAN, (3) difficulties in separating PCs from the surrounding myocardium, (4) lack of specific molecular markers, and (5) significant cell heterogeneity in SAN tissues [10][11].
The application of single-cell RNA sequencing (scRNA-seq) technology has enabled the investigation of gene regulation, cell diversification and temporality, cellular heterogeneity, and developmental temporality from a global and unbiased perspective with unprecedented resolution [12]. The scRNA-seq technology can overcome several obstacles in SAN research, including low cell numbers, complex and variable structures, distorted transcriptional profiles due to cell heterogeneity, and nonconductive cell contamination [10].
2. SAN Structure
The human SAN is a compact, slightly elongated, banana-shaped structure located at the junction of the lateral superior vena cava (SVC) and right atrium (RA). Functional and structural mapping of the human SAN has revealed that it can be distinguished into head, center, and tail compartments. The head is usually closer to the epicardium, and the tail is typically tilted toward the endocardium [13][14][15][16][17]. Different regions of the SAN pacemaker clusters exhibit diverse functional and molecular features, which are activated through distinct cellular mechanisms [18][19][20] (Figure 1A).
In mice, the SAN artery, autonomic nerve fibers, telocytes, monocytes, macrophages, adipocytes, and fibroblasts were observed via electron microscopy [21]. High populations of fibroblasts in the SAN maintain the source–sink balance and ensure normal electrical conduction [3]. The heterogeneous structure of the SAN makes studying its function challenging. Development of the scRNA-seq technology has aided in examining these heterogeneous and rare cells.
2.1. Three-Dimensional Structure of the SAN
In adult mice, the SAN head/center, which is densely packed with clusters of PCs, is the leading pacemaker region, extends superiorly, and wraps the right SVC [22]. The SAN also extends inferiorly toward the inferior vena cava (IVC), forming the tail [23][24]. In mice, the SAN can be genetically divided into a TBX18+/NKX2-5− head domain and a TBX18−/NKX2-5+ tail domain [7] (Figure 1B,C). TBX18 deletion in the SAN head of mice has been shown to result in a normal heart rate [7], whereas SHOX2 ablation in the SAN tail has led to severe SSS [25]. These findings highlight the different roles of different parts of the SAN in pacemaking and conduction.
Understanding the heterogeneity of the SAN complex is necessary to fully characterize its pacing and conduction functions. To further explore the complex 3D anatomy of the SAN, Goodyer et al. used scRNA-seq in E16.5 mice [10]. The cells underwent unsupervised clustering via t-distributed stochastic neighbor embedding (t-SNE). Cluster 9 showed significant enrichment of nodal markers. Further experiments identified two distinct subclusters within cluster 9, i.e., compact SAN (cSAN) and transitional cells (Tz). cSAN showed high levels of established SAN markers with low or no known atrial myocardium (AM) marker expression [10]. Further investigation of the cellular heterogeneity of the cSAN by Goodyer et al. revealed two distinct clusters in the cSAN, consistent with the previously recognized head and tail subdomains [10]. Both clusters showed high expressions of the nodal markers HCN4 and SHOX2. The head cluster showed increased expression of TBX18 and decreased expression of NKX2-5, whereas the tail cluster showed decreased expression of TBX18 and increased expression of NKX2-5 [7], consistent with previous reports [26]. A comparison of these two clusters revealed differentially expressed genes not previously reported. IGFBP5, PDE1A, and VSNL1 were abundant in the head, whereas SMPX, ALDH1B1, and SLC22A1 were abundant in the tail. These findings may enable the examination of different functional characteristics of the head and tail [10].
To further explore the different functional and molecular bases of the SAN head and tail, Li et al. (2019) examined the action potentials (APs) of the SAN’s head and tail separately using a patch clamp and found that the SAN tail has unique APs that mediate the SAN head and A. By performing scRNA-seq assays on SAN regions highlighted by GFP isolated from E13.5 SHOX2Cre/+ in R26RmTmG mice, they also verified that NKX2-5 plays an important role in the development and function of the SAN tail. They found four cardiomyocyte subpopulations, one of which expressed both SAN- and AM-specific genes, consistent with the properties of the SAN tail. They also revealed that the SAN tail has unique electrophysiological properties and transcriptomic features [27].
According to Li et al.’s integrated NIOM-3D structural study, the human SAN is functionally insulated/discontinued from the atria by fibrotic tissue, fat, and myofibers, with the exception of 2–5 discrete sinoatrial conduction pathways (SACPs) [17][28]. SACPs transmit electrical impulses to the RA [29][30]. SACP structure includes tracts of myofibers with transitional cells on the SAN border that merge with atrial myobundles [31].
The human fetal SAN contains predominantly PCs and limited collagen and transitional cells. From approximately 2 weeks after birth, Tz gradually increase in number and intertwine with the collagen framework [32]. Importantly, Tz have been implicated in SSS [31]. However, they remain poorly understood due to identification and isolation challenges. Goodyer et al. provided transcriptomic data for Tz [10]. In their study, Tz expressed lower levels of nodal markers and were enriched for AM markers GJA5 and SCN5A [10]. However, no new markers were specific to Tz, and it remains uncertain as to how Tz enter the SAN architecture.
2.2. SAN Microenvironment
The unique microenvironment of the SAN maintains its functional characteristics. However, the formation and function of the SAN microenvironment remain poorly understood. To determine the cellular processes that contribute to the formation of the SAN microenvironment, Bressan et al. performed RNA-seq on chicken embryos earlier than E3.5 and found that factors associated with epithelial-to-mesenchymal transition and fibrosis were upregulated in the pacemaker region [33]. Whole-mount in situ hybridization and microsurgery demonstrated that proepicardium-derived mesenchymal cells (including fibroblasts) contributed to the formation of the SAN microenvironment and that the integration of mesenchymal cells is essential to protecting PCs from the electrical “load” of the adjacent AM [33]. However, how mesenchymal cells interact with PCs and regulate physiological functions in the SAN requires further investigation.
Recently, Chou et al. compared TBX18-induced PCs and ventricular myocardium via RNA-seq and showed the predominance of glucose metabolism and glycolysis in the GO analysis [34]. TBX18-induced PCs co-culturing with fibroblasts activated the integrin-dependent mitogen-activated protein kinase-E2F1 signal through cell–cell contact and induced Aldoc expression in PCs [34]. Their results provided new insights into the link between fibroblasts and PCs and a new way to mediate SSS therapy by regulating Aldoc.
The human SAN comprises 35%–55% fibrotic tissue [35]; cardiac diseases may further increase fibrosis within the SAN, potentially impairing automaticity and the conduction of electricity to the atria. Recently, Li et al. described the transcriptional characteristics of fibroblasts in the SANs of patients with heart failure but did not further elaborate on the connection between fibroblasts and PCs [36].
In addition to fibroblasts, many other cell types exist in the SAN [21]. The interactions among these cells and their roles in maintaining sinus rhythm have not been well elucidated. Single-cell transcriptomics is a promising tool for investigating cell–cell interaction networks [37], which is also the direction of the research group in the future.
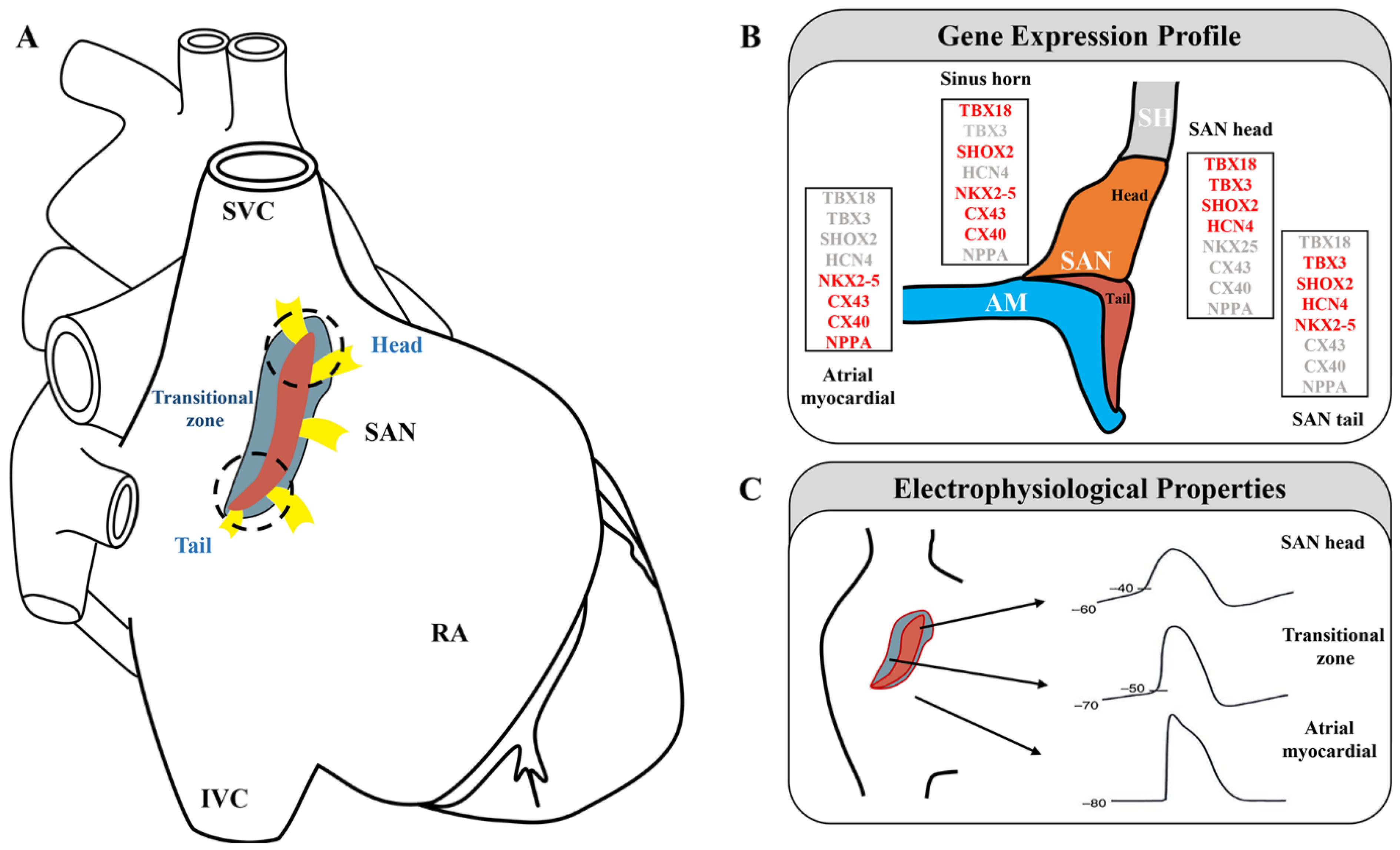
Figure 1. Sinoatrial node subdomains are divided based on anatomical and genetic criteria. (A) The sinoatrial node is displayed on a schematic diagram of the heart (red, central SAN region; blue, transitional cells; yellow, sinoatrial conduction pathways; white dashed line, SAN head; black dashed line, SAN tail). Adapted with permission from [24][28], Copyright (2005, 2017). (B) Gene expression profile in the SAN region (expressed genes are indicated in red). Adapted with permission from [7], Copyright (2009). (C) Samples of action potentials (APs) are recorded from the central sinoatrial node to the periphery of the node. Adapted with permission from [20], Copyright (2016).
3. Transcriptional Regulation and Specific Molecular Markers of the SAN
SAN formation and function are tightly regulated by a TF network that displays a dynamic and unique expression pattern in the SAN and surrounding AM [38]. These TFs are key players in the development and differentiation of PCs and help maintain pacemaker identity and function. A previous study on SAN formation and development used a gene-deficient mouse model; their findings revealed important aspects of SAN development and differentiation [4][5][6][7][8] (Figure 2). However, how key developmental regulators are regulated in individual cells at specific locations during the SAN development remains unclear.
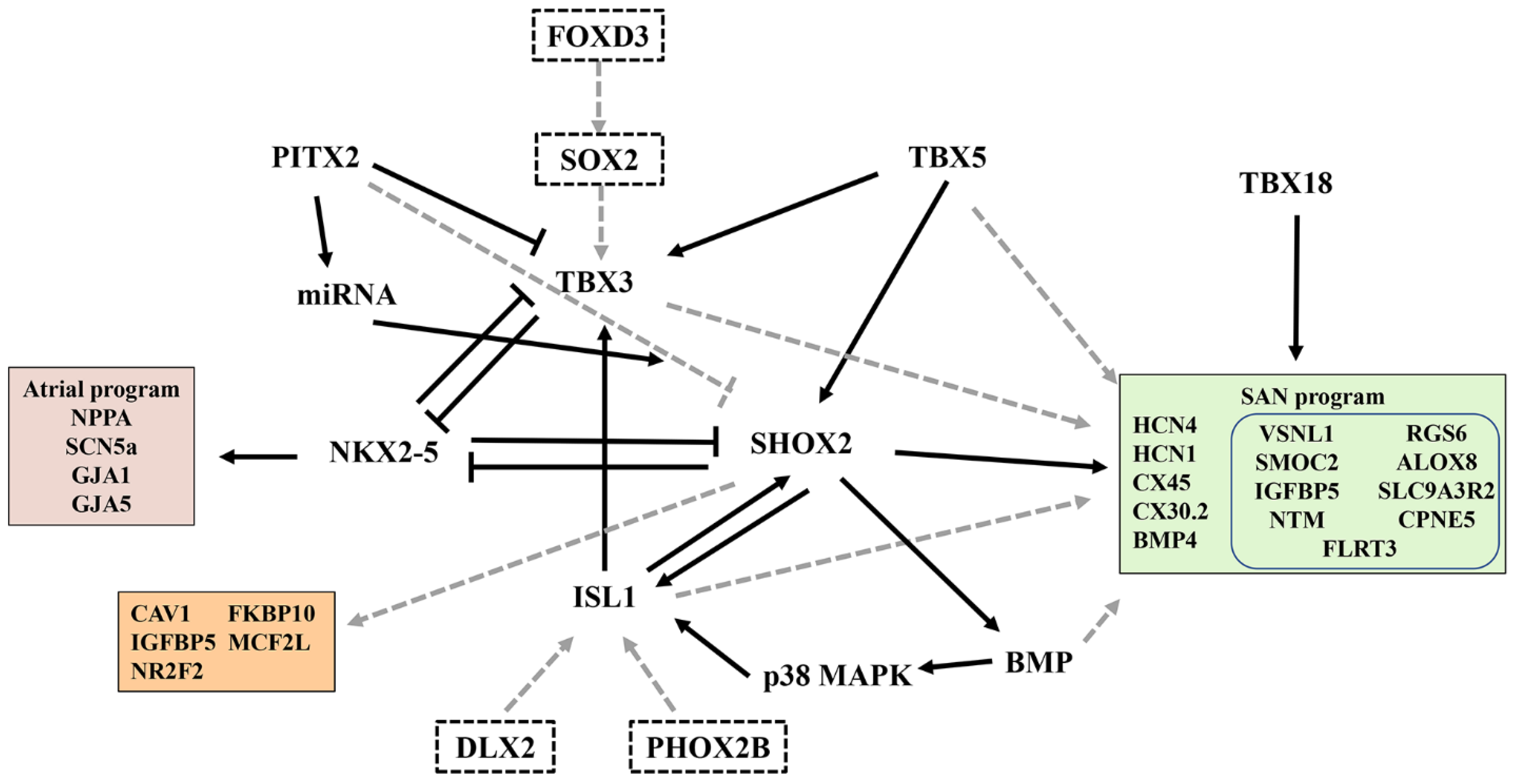
Figure 2. A gene regulatory network that controls SAN development and function. The black arrows indicate gene activation. Black lines with blunt ends represent the inhibition of gene expression by their corresponding transcription factors. The dotted lines indicate indirect or speculation effects. The right box represents the SAN program, and the blue box indicates the SAN-related gene discovered by single-cell sequencing. The left upper box represents the atrial program; the left lower box indicates the genes regulated by SHOX2. The dotted box indicates the potential transcription factors associated with the SAN.
One of the characteristics of the SAN is the high expression of the hyperpolarization-activated and cyclic-nucleotide-gated ion channel HCN4 [39], which underlies the funny current (If), an essential factor for the maintenance of sinus rhythm [40]. In humans, HCN4 is expressed in PCs and RA, and so cannot be used for identifying human PCs as the sole marker [41]. CD166/Alcam, a cell surface molecule, can help identify CD166+ pacemaker precursors from differentiating mouse embryonic stem cells. However, CD166/Alcam is not expressed in PCs; thus, its application is limited [42][43]. Therefore, specific cell surface molecules of PCs are still being sought.
3.1. Transcription Factors
Recently, the critical regulatory role of ISL1 in SAN development has received increasing attention [44][45]. The specific role of ISL1 in the SAN is unknown because of the early death of ISL1-deficient mice and the loss of cardiomyocytes derived from ISL1 progenitor cells [38]. Recent transcriptome studies have shown significant changes in SAN-related genes when ISL1 is ablated in the SAN [44]. ISL1 deficiency in mice leads to the downregulation of TBX3, SHOX2, and BMP4, which are key regulators of SAN development, as well as HCN4, HCN1, and CACNA1G, which are responsible for the functioning of ion channels of the SAN [44]. However, expressions of NPPA, PITX2, NKX2.5, GJA1, GJA5, and SCN5A were upregulated. Their study highlighted the central role of ISL1 in establishing the PC gene program [44].
SHOX2 plays a key role in PC fate and can activate the pacemaker gene program (TBX3, ISL1, and HCN4) by repressing NKX2-5 and the working myocardial gene program [38]. Hoffmann et al. performed transcriptome profiling of SHOX2+/+ vs. SHOX2−/− ESC-derived SANLPCs via RNA-seq to explore SHOX2 pathways involved in pacemaker differentiation [46]. Interestingly, some SAN-related genes (TBX3, ISL1, HCN4, and NKX2-5) were not affected by SHOX2 deletion. On combining this finding with data obtained by Vedantham et al. [44], several SHOX2 target genes were discovered, namely, CAV1, FKBP10, IGFBP5, MCF2l, and NR2F2, which were validated in mouse and zebrafish models [46]. Their study provided new insights into the transcriptional regulation of SHOX2 during pacemaker development and function (Figure 2).
The homeobox transcription factor PITX2 is a laterality gene responsible for establishing the right- and left-body axes, asymmetric gene expression, and organ morphogenesis [47]. Bilateral or ectopic SAN can be found in PITX2-deficient embryos and may contribute to atrial fibrillation in adult animals with reduced PITX2 expression [48][49]. Single-cell transcriptomics has revealed the role of PITX2 in cardiac development and left–right cellular specification [50]. The CM-RA1 cluster with a SAN transcriptional signature was more abundant in PITX2 mutants than that in controls [50], highlighting the inhibitory effect of PITX2 on SAN development.
In addition to the above TFs that affect SAN development and function, next-generation sequencing was recently used to identify other unexplored novel TFs in human SAN and RA and to predict interactions between key TFs and genes involved in pacemaker mechanisms [51]. In the adult SAN, many new TFs were highly expressed (e.g., FOXD3, DLX2, PHOX2B, VENTX, and SOX2). However, the role of these TFs in the adult mammalian SAN is unknown and should be explored in the future (Figure 2).
A unique set of TFs, which are enriched in PCs, act as activators and repressors and interact with each other to determine the fate of the SAN [38]. Some TFs have been used to direct stem cell differentiation toward SANLPCs [52]; however, the core TF set requires further exploration to obtain highly pure SANLPCs.
3.2. Specific Molecular Markers of the SAN
In addition to the enrichment of established nodal genes, scRNA-seq revealed a host of significant novel genes not previously reported to be involved in SAN development or function, including insulin growth factor binding protein 5 (IGFBP5) [10], SPARC-related modular calcium-binding protein 2 (SMOC2) [10][53], neurotrimin (NTM) [10], copine 5 (CPNE5) [10], regulator of G-protein signaling type 6 (RGS6) [10][54], arachidonate 8-lipoxygenase (ALOX8) [53], sodium–hydrogen exchange regulatory cofactor 2 (SLC9A3R2) [53], and fibronectin leucine-rich transmembrane protein 3 (FLRT3) [55] (Figure 2).
Van Eif et al. used CRISPR/Cas9 to produce SMOC2 frameshift mutation mice and verify the effect of SMOC2 on SAN function. In vivo and in vitro experiments revealed that SMOC2 inactivation had little effect on cardiac electrophysiology [53]. Interestingly, SMOC2 has recently been reported as a new SAN marker. The gene was subsequently validated via immunostaining or fluorescence in situ hybridization, showing SMOC2 enrichment within the SAN compared with the surrounding AM [10]. In a recent study, scRNA-seq of single cells from the SAN of different mammals identified a species-conserved potential SAN marker, VSNL1 (a member of the visinin/recoverin subfamily of neuronal calcium sensor proteins), which is abundantly expressed in the SAN but barely expressed in the AM or ventricles [56]. Although its function is unknown, VSNL1 expression has been detected in the venous pole region of the developing heart [57]. Moreover, VSNL1 deficiency reduces the heart rate in human-induced pluripotent stem cell-derived cardiomyocytes and mice [56]. FLRT3, a cell-autonomous regulator of the adherens junction of PCs, can mediate the transmission of electrical activity by regulating gap junctions [55].
References
- Wallace, M.J.; El Refaey, M.; Mesirca, P.; Hund, T.J.; Mangoni, M.E.; Mohler, P.J. Genetic Complexity of Sinoatrial Node Dysfunction. Front. Genet. 2021, 12, 654925.
- Weiss, J.N.; Qu, Z. The Sinus Node: Still Mysterious After All These Years. JACC Clin. Electrophysiol. 2020, 6, 1841–1843.
- Csepe, T.A.; Kalyanasundaram, A.; Hansen, B.J.; Zhao, J.; Fedorov, V.V. Fibrosis: A structural modulator of sinoatrial node physiology and dysfunction. Front Physiol 2015, 6, 37.
- Mori, A.D.; Zhu, Y.; Vahora, I.; Nieman, B.; Koshiba-Takeuchi, K.; Davidson, L.; Pizard, A.; Seidman, J.G.; Seidman, C.E.; Chen, X.J.; et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 2006, 297, 566–586.
- Hoogaars, W.M.; Engel, A.; Brons, J.F.; Verkerk, A.O.; de Lange, F.J.; Wong, L.Y.; Bakker, M.L.; Clout, D.E.; Wakker, V.; Barnett, P.; et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007, 21, 1098–1112.
- Espinoza-Lewis, R.A.; Yu, L.; He, F.; Liu, H.; Tang, R.; Shi, J.; Sun, X.; Martin, J.F.; Wang, D.; Yang, J.; et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009, 327, 376–385.
- Wiese, C.; Grieskamp, T.; Airik, R.; Mommersteeg, M.T.; Gardiwal, A.; de Gier-de Vries, C.; Schuster-Gossler, K.; Moorman, A.F.; Kispert, A.; Christoffels, V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 2009, 104, 388–397.
- Mommersteeg, M.T.; Hoogaars, W.M.; Prall, O.W.; de Gier-de Vries, C.; Wiese, C.; Clout, D.E.; Papaioannou, V.E.; Brown, N.A.; Harvey, R.P.; Moorman, A.F.; et al. Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 2007, 100, 354–362.
- Mantri, S.; Wu, S.M.; Goodyer, W.R. Molecular Profiling of the Cardiac Conduction System: The Dawn of a New Era. Curr. Cardiol. Rep. 2021, 23, 103.
- Goodyer, W.R.; Beyersdorf, B.M.; Paik, D.T.; Tian, L.; Li, G.; Buikema, J.W.; Chirikian, O.; Choi, S.; Venkatraman, S.; Adams, E.L.; et al. Transcriptomic Profiling of the Developing Cardiac Conduction System at Single-Cell Resolution. Circ. Res. 2019, 125, 379–397.
- Park, D.S.; Fishman, G.I. The cardiac conduction system. Circulation 2011, 123, 904–915.
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502.
- Sanchez-Quintana, D.; Cabrera, J.A.; Farre, J.; Climent, V.; Anderson, R.H.; Ho, S.Y. Sinus node revisited in the era of electroanatomical mapping and catheter ablation. Heart 2005, 91, 189–194.
- Keith, A.; Flack, M. The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J. Anat. Physiol. 1907, 41, 172–189.
- James, T.N.; Sherf, L.; Fine, G.; Morales, A.R. Comparative ultrastructure of the sinus node in man and dog. Circulation 1966, 34, 139–163.
- Truex, R.C.; Smythe, M.Q.; Taylor, M.J. Reconstruction of the human sinoatrial node. Anat. Rec. 1967, 159, 371–378.
- Kalyanasundaram, A.; Li, N.; Augostini, R.S.; Weiss, R.; Hummel, J.D.; Fedorov, V.V. Three-Dimensional Functional Anatomy of Human Sinoatrial node for Epicardial and Endocardial Mapping and Ablation. Heart Rhythm. 2022, 22, S1547–S5271.
- Lang, D.; Petrov, V.; Lou, Q.; Osipov, G.; Efimov, I.R. Spatiotemporal control of heart rate in a rabbit heart. J. Electrocardiol. 2011, 44, 626–634.
- Monfredi, O.; Tsutsui, K.; Ziman, B.; Stern, M.D.; Lakatta, E.G.; Maltsev, V.A. Electrophysiological heterogeneity of pacemaker cells in the rabbit intercaval region, including the SA node: Insights from recording multiple ion currents in each cell. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H403–H414.
- Murphy, C.; Lazzara, R. Current concepts of anatomy and electrophysiology of the sinus node. J. Interv. Card. Electrophysiol. 2016, 46, 9–18.
- Linscheid, N.; Logantha, S.; Poulsen, P.C.; Zhang, S.; Schrolkamp, M.; Egerod, K.L.; Thompson, J.J.; Kitmitto, A.; Galli, G.; Humphries, M.J.; et al. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat. Commun. 2019, 10, 2889.
- Verheijck, E.E.; van Kempen, M.J.; Veereschild, M.; Lurvink, J.; Jongsma, H.J.; Bouman, L.N. Electrophysiological features of the mouse sinoatrial node in relation to connexin distribution. Cardiovasc. Res. 2001, 52, 40–50.
- Dobrzynski, H.; Boyett, M.R.; Anderson, R.H. New insights into pacemaker activity: Promoting understanding of sick sinus syndrome. Circulation 2007, 115, 1921–1932.
- Dobrzynski, H.; Li, J.; Tellez, J.; Greener, I.D.; Nikolski, V.P.; Wright, S.E.; Parson, S.H.; Jones, S.A.; Lancaster, M.K.; Yamamoto, M.; et al. Computer three-dimensional reconstruction of the sinoatrial node. Circulation 2005, 111, 846–854.
- Ye, W.; Wang, J.; Song, Y.; Yu, D.; Sun, C.; Liu, C.; Chen, F.; Zhang, Y.; Wang, F.; Harvey, R.P.; et al. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 2015, 142, 2521–2532.
- Christoffels, V.M.; Smits, G.J.; Kispert, A.; Moorman, A.F. Development of the pacemaker tissues of the heart. Circ. Res. 2010, 106, 240–254.
- Li, H.; Li, D.; Wang, Y.; Huang, Z.; Xu, J.; Yang, T.; Wang, L.; Tang, Q.; Cai, C.L.; Huang, H.; et al. Nkx2-5 defines a subpopulation of pacemaker cells and is essential for the physiological function of the sinoatrial node in mice. Development 2019, 146, dev178145.
- Li, N.; Hansen, B.J.; Csepe, T.A.; Zhao, J.; Ignozzi, A.J.; Sul, L.V.; Zakharkin, S.O.; Kalyanasundaram, A.; Davis, J.P.; Biesiadecki, B.J.; et al. Redundant and diverse intranodal pacemakers and conduction pathways protect the human sinoatrial node from failure. Sci. Transl. Med. 2017, 9, eaam5607.
- Boineau, J.P.; Canavan, T.E.; Schuessler, R.B.; Cain, M.E.; Corr, P.B.; Cox, J.L. Demonstration of a widely distributed atrial pacemaker complex in the human heart. Circulation 1988, 77, 1221–1237.
- Fedorov, V.V.; Glukhov, A.V.; Chang, R. Conduction barriers and pathways of the sinoatrial pacemaker complex: Their role in normal rhythm and atrial arrhythmias. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H1773–H1783.
- Csepe, T.A.; Zhao, J.; Hansen, B.J.; Li, N.; Sul, L.V.; Lim, P.; Wang, Y.; Simonetti, O.P.; Kilic, A.; Mohler, P.J.; et al. Human sinoatrial node structure: 3D microanatomy of sinoatrial conduction pathways. Prog. Biophys. Mol. Biol. 2016, 120, 164–178.
- James, T.N. Structure and function of the sinus node, AV node and His bundle of the human heart: Part I-structure. Prog. Cardiovasc. Dis. 2002, 45, 235–267.
- Bressan, M.; Henley, T.; Louie, J.D.; Liu, G.; Christodoulou, D.; Bai, X.; Taylor, J.; Seidman, C.E.; Seidman, J.G.; Mikawa, T. Dynamic Cellular Integration Drives Functional Assembly of the Heart’s Pacemaker Complex. Cell Rep. 2018, 23, 2283–2291.
- Chou, P.C.; Liu, C.M.; Weng, C.H.; Yang, K.C.; Cheng, M.L.; Lin, Y.C.; Yang, R.B.; Shyu, B.C.; Shyue, S.K.; Liu, J.D.; et al. Fibroblasts Drive Metabolic Reprogramming in Pacemaker Cardiomyocytes. Circ. Res. 2022, 131, 6–20.
- Shiraishi, I.; Takamatsu, T.; Minamikawa, T.; Onouchi, Z.; Fujita, S. Quantitative histological analysis of the human sinoatrial node during growth and aging. Circulation 1992, 85, 2176–2184.
- Kalyanasundaram, A.; Li, N.; Gardner, M.L.; Artiga, E.J.; Hansen, B.J.; Webb, A.; Freitas, M.A.; Pietrzak, M.; Whitson, B.A.; Mokadam, N.A.; et al. Fibroblast-Specific Proteo-Transcriptomes Reveal Distinct Fibrotic Signatures of Human Sinoatrial Node in Non-Failing and Failing Hearts. Circulation 2021, 144, 126–143.
- Zhang, L.; Li, Z.; Skrzypczynska, K.M.; Fang, Q.; Zhang, W.; O’Brien, S.A.; He, Y.; Wang, L.; Zhang, Q.; Kim, A.; et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 2020, 181, 442–459.e429.
- Van Eif, V.W.W.; Devalla, H.D.; Boink, G.J.J.; Christoffels, V.M. Transcriptional regulation of the cardiac conduction system. Nat. Rev. Cardiol. 2018, 15, 617–630.
- Chandler, N.J.; Greener, I.D.; Tellez, J.O.; Inada, S.; Musa, H.; Molenaar, P.; Difrancesco, D.; Baruscotti, M.; Longhi, R.; Anderson, R.H.; et al. Molecular architecture of the human sinus node: Insights into the function of the cardiac pacemaker. Circulation 2009, 119, 1562–1575.
- Darche, F.F.; Rivinius, R.; Kollensperger, E.; Leimer, U.; Germann, G.; Seckinger, A.; Hose, D.; Schroter, J.; Bruehl, C.; Draguhn, A.; et al. Pacemaker cell characteristics of differentiated and HCN4-transduced human mesenchymal stem cells. Life Sci. 2019, 232, 116620.
- Kalyanasundaram, A.; Li, N.; Hansen, B.J.; Zhao, J.; Fedorov, V.V. Canine and human sinoatrial node: Differences and similarities in the structure, function, molecular profiles, and arrhythmia. J. Vet. Cardiol. 2019, 22, 2–19.
- Scavone, A.; Capilupo, D.; Mazzocchi, N.; Crespi, A.; Zoia, S.; Campostrini, G.; Bucchi, A.; Milanesi, R.; Baruscotti, M.; Benedetti, S.; et al. Embryonic stem cell-derived CD166+ precursors develop into fully functional sinoatrial-like cells. Circ. Res. 2013, 113, 389–398.
- Liang, X.; Evans, S.M.; Sun, Y. Insights into cardiac conduction system formation provided by HCN4 expression. Trends Cardiovasc. Med. 2015, 25, 1–9.
- Vedantham, V.; Galang, G.; Evangelista, M.; Deo, R.C.; Srivastava, D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ. Res. 2015, 116, 797–803.
- Liang, X.; Zhang, Q.; Cattaneo, P.; Zhuang, S.; Gong, X.; Spann, N.J.; Jiang, C.; Cao, X.; Zhao, X.; Zhang, X.; et al. Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Investig. 2015, 125, 3256–3268.
- Hoffmann, S.; Schmitteckert, S.; Raedecke, K.; Rheinert, D.; Diebold, S.; Roeth, R.; Weiss, B.; Granzow, M.; Niesler, B.; Griesbeck, A.; et al. Network-driven discovery yields new insight into Shox2-dependent cardiac rhythm control. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194702.
- Galli, D.; Dominguez, J.N.; Zaffran, S.; Munk, A.; Brown, N.A.; Buckingham, M.E. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development 2008, 135, 1157–1167.
- Ammirabile, G.; Tessari, A.; Pignataro, V.; Szumska, D.; Sutera Sardo, F.; Benes, J., Jr.; Balistreri, M.; Bhattacharya, S.; Sedmera, D.; Campione, M. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 2012, 93, 291–301.
- Wang, J.; Klysik, E.; Sood, S.; Johnson, R.L.; Wehrens, X.H.; Martin, J.F. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. USA 2010, 107, 9753–9758.
- Hill, M.C.; Kadow, Z.A.; Li, L.; Tran, T.T.; Wythe, J.D.; Martin, J.F. A cellular atlas of Pitx2-dependent cardiac development. Development 2019, 146, dev180398.
- Aminu, A.J.; Petkova, M.; Atkinson, A.J.; Yanni, J.; Morris, A.D.; Simms, R.T.; Chen, W.; Yin, Z.; Kuniewicz, M.; Holda, M.K.; et al. Further insights into the molecular complexity of the human sinus node—The role of ‘novel’ transcription factors and microRNAs. Prog. Biophys. Mol. Biol. 2021, 166, 86–104.
- Raghunathan, S.; Islas, J.F.; Mistretta, B.; Iyer, D.; Shi, L.; Gunaratne, P.H.; Ko, G.; Schwartz, R.J.; McConnell, B.K. Conversion of human cardiac progenitor cells into cardiac pacemaker-like cells. J. Mol. Cell. Cardiol. 2020, 138, 12–22.
- Van Eif, V.W.W.; Stefanovic, S.; van Duijvenboden, K.; Bakker, M.; Wakker, V.; de Gier-de Vries, C.; Zaffran, S.; Verkerk, A.O.; Boukens, B.J.; Christoffels, V.M. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development 2019, 146, dev173161.
- Yang, J.; Huang, J.; Maity, B.; Gao, Z.; Lorca, R.A.; Gudmundsson, H.; Li, J.; Stewart, A.; Swaminathan, P.D.; Ibeawuchi, S.R.; et al. RGS6, a modulator of parasympathetic activation in heart. Circ. Res. 2010, 107, 1345–1349.
- Thomas, K.; Henley, T.; Rossi, S.; Costello, M.J.; Polacheck, W.; Griffith, B.E.; Bressan, M. Adherens junction engagement regulates functional patterning of the cardiac pacemaker cell lineage. Dev. Cell 2021, 56, 1498–1511.e7.
- Liang, D.; Xue, J.; Geng, L.; Zhou, L.; Lv, B.; Zeng, Q.; Xiong, K.; Zhou, H.; Xie, D.; Zhang, F.; et al. Cellular and molecular landscape of mammalian sinoatrial node revealed by single-cell RNA sequencing. Nat. Commun. 2021, 12, 287.
- Ola, R.; Lefebvre, S.; Braunewell, K.H.; Sainio, K.; Sariola, H. The expression of Visinin-like 1 during mouse embryonic development. Gene Expr. Patterns 2012, 12, 53–62.
More
Information
Subjects:
Developmental Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.4K
Revisions:
2 times
(View History)
Update Date:
09 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




