
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stephen Gaunt | -- | 2573 | 2022-12-01 11:49:10 | | | |
| 2 | Stephen Gaunt | + 276 word(s) | 2920 | 2022-12-15 12:51:40 | | | | |
| 3 | Camila Xu | + 346 word(s) | 2921 | 2022-12-19 07:34:28 | | | | |
| 4 | Stephen Gaunt | -1 word(s) | 2921 | 2022-12-20 12:13:04 | | |
Video Upload Options
The Hox gene cluster, responsible for patterning of the head–tail axis, is an ancestral feature of all bilaterally symmetrical animals (the Bilateria) that remains intact in a wide range of species.
1. Introduction
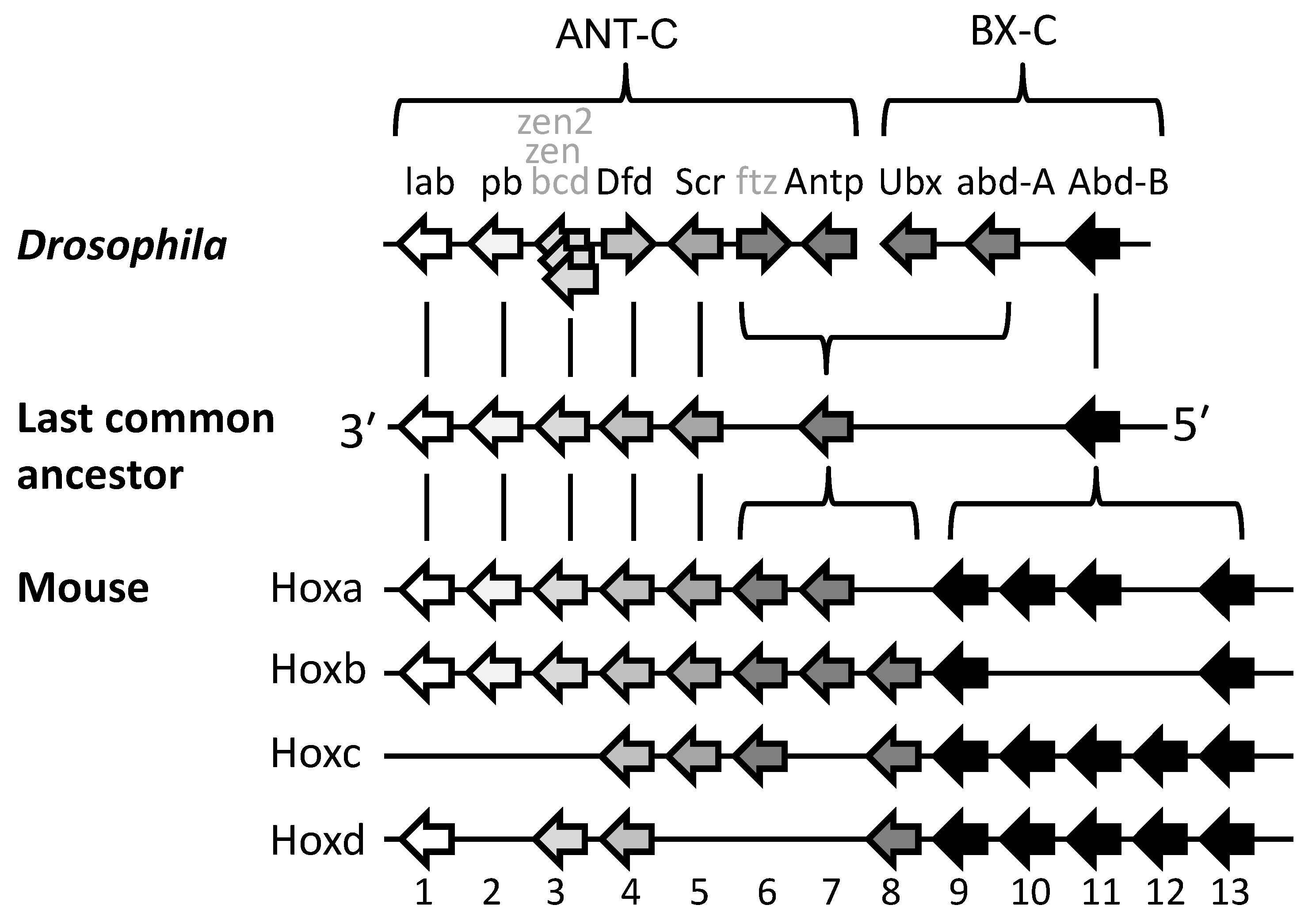
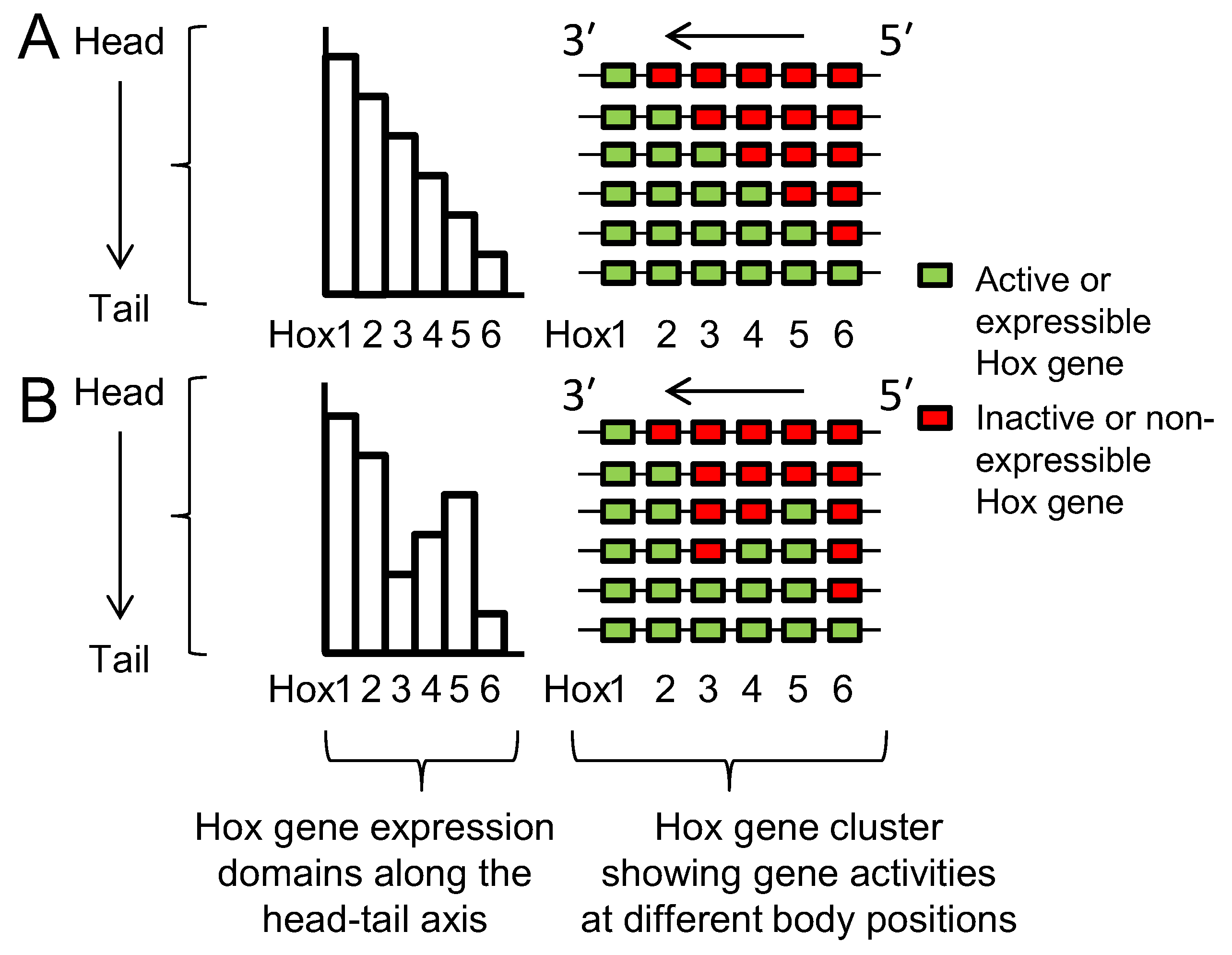
Lewis’s model describes Hox genes as expressing or non-expressing, but researchers now understand that these states are more accurately described as, respectively, expressible or non-expressible. Recent studies in both mice [9][10] and Drosophila [11] show that the expressible Hox genes in a given cell (Figure 2A-right) are in an open chromatin state, characterized by Trithorax (Trx) protein binding, while the non-expressible genes are in a closed chromatin state, characterized by Polycomb (Pc) protein binding (Figure 3). These chromatin states are usually heritable from one cell generation to the next, thereby ensuring that Hox expressibility patterns acquired in the early embryo are faithfully remembered at all later stages, enabling guidance throughout the course of development. At any region along the body, there is typically only one boundary between expressible and non-expressible genes in both mice [9][10] (Figure 2A-right and Figure 3) and Drosophila [11]. In the subtle difference from Lewis’s original proposal, expressible genes need not always be expressed. Expression depends upon the availability of activating or repressive transcription factors and may change in a tissue over developmental time. Expressible Hox genes have been described as “open for business” [12][13]. Regions of the body where Hox genes are non-expressible (closed for business; red zones in Figure 3) typically cannot express these genes at any time.
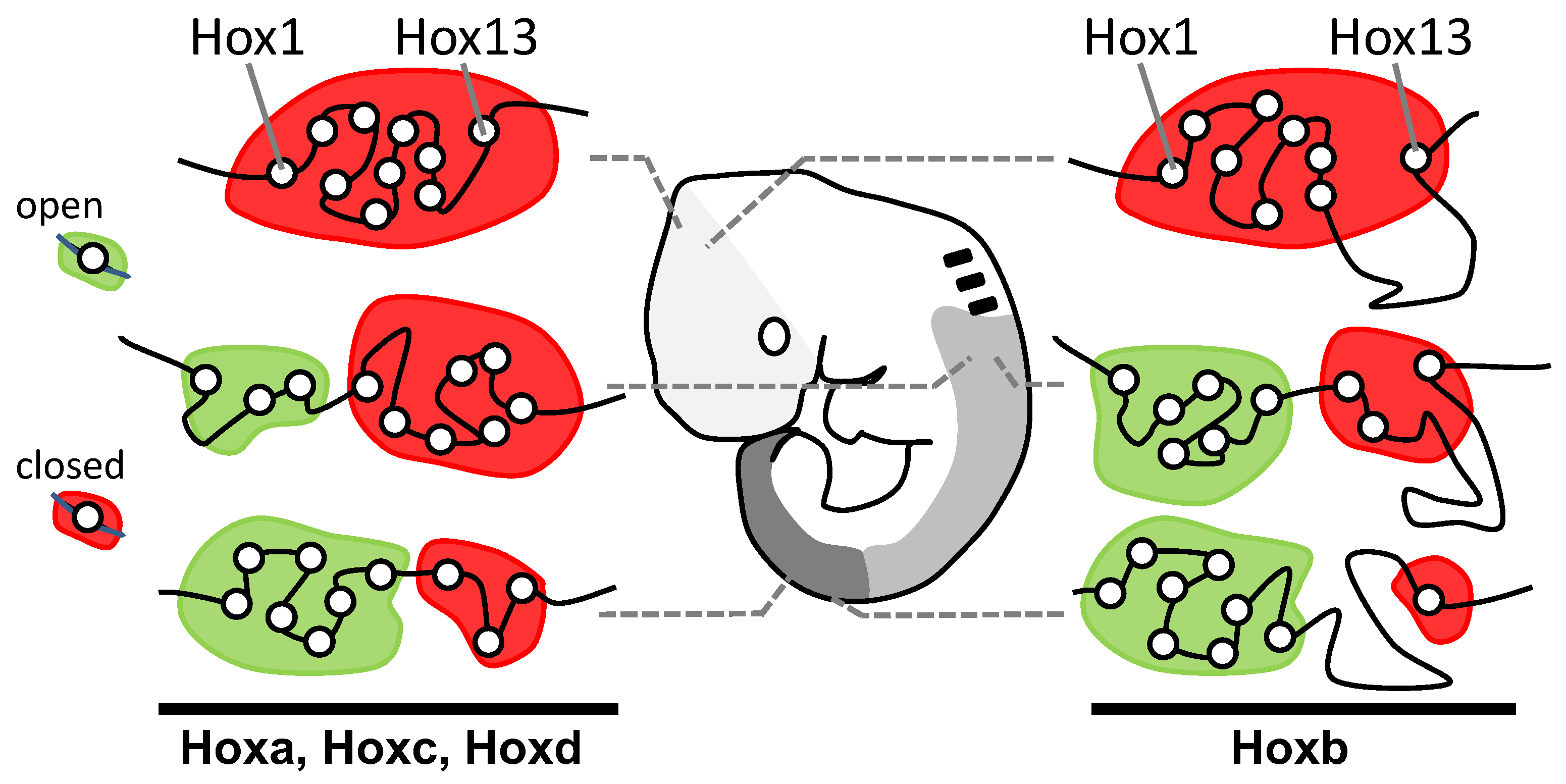
Figure 3. Discreet domains of open and closed chromatin generally support Lewis’s model. Antibody studies show correspondence between the position of cells along the head–tail axis and the distributions of Hox genes between open (Hox expressible, green domain; Trithorax-rich) and closed (Hox non-expressible, red domain; Polycomb-rich) chromatin states. At each level along the body, there is only a single boundary between these states, supporting Lewis’s model (Figure 2A-right). Re-drawn/modified from Noordermeer et al. [10].
2. Seeking Sense in the Hox Gene Cluster
2.1. Seeking Sense in the Evolutionary Origin of Hox Clustering and Transcriptional Direction
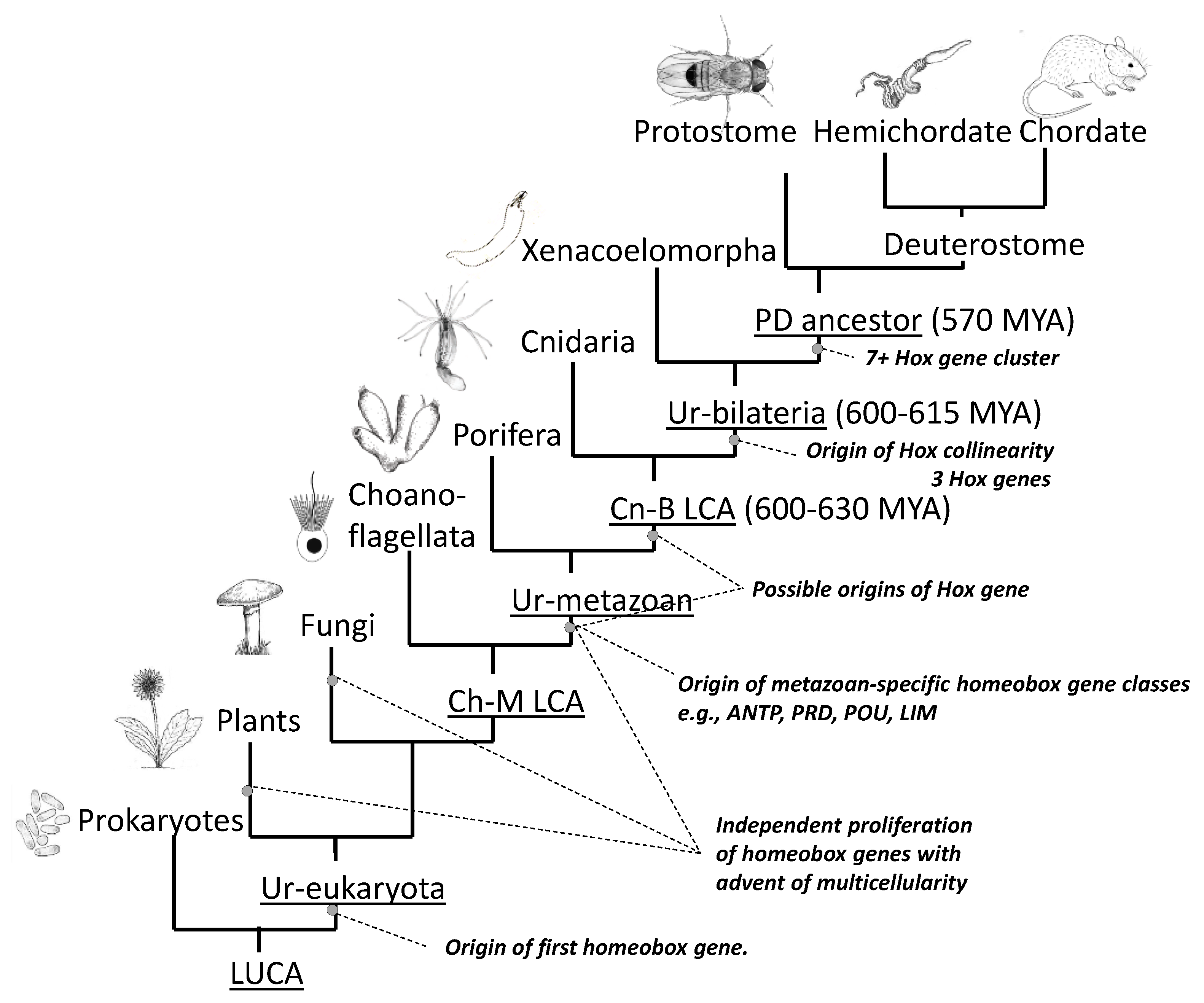
2.2. Seeking Sense in the Maintenance and Compactness of Clusters
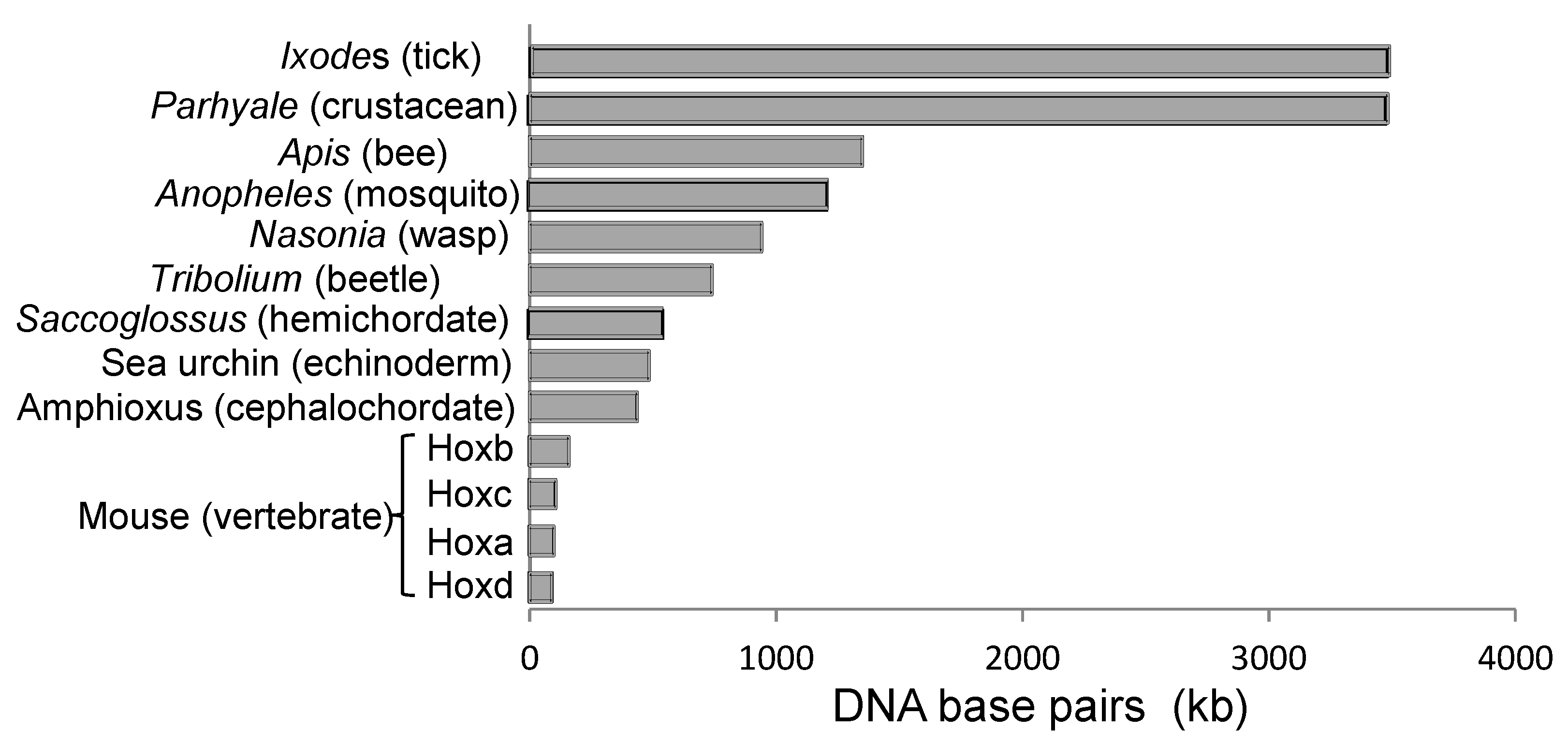
2.3. Seeking Sense in Spatial and Temporal Collinearities
References
- Boncinelli, E.; Somma, R.; Acampora, D.; Pannese, M.; D’Esposito, M.; Faiella, A.; Simeone, A. Organization of human homeobox genes. Hum. Reprod. 1988, 3, 880–886.
- Duboule, D.; Dolle, P. The structural and functional organization of the murine hox gene family resembles that of drosophila homeotic genes. EMBO J. 1989, 8, 1497–1505.
- Graham, A.; Papalopulu, N.; Krumlauf, R. The murine and drosophila homeobox gene complexes have common features of organization and expression. Cell 1989, 57, 367–378.
- Balavoine, G.; de Rosa, R.; Adoutte, A. Hox clusters and bilaterian phylogeny. Mol. Phylogenet. Evol. 2002, 24, 366–373.
- Gaunt, S.J. Hox cluster genes and collinearities throughout the tree of animal life. Int. J. Dev. Biol. 2018, 62, 673–683.
- Gaunt, S.J. Made in the Image of a Fly; Amazon Books and Ebooks, 2019.
- Lewis, E.B. A gene complex controlling segmentation in drosophila. Nature 1978, 276, 565–570.
- Gaunt, S.J. The significance of hox gene collinearity. Int. J. Dev. Biol. 2015, 59, 159–170.
- Soshnikova, N.; Duboule, D. Epigenetic temporal control of mouse hox genes in vivo. Science 2009, 324, 1320–1323.
- Noordermeer, D.; Leleu, M.; Splinter, E.; Rougemont, J.; De Laat, W.; Duboule, D. The dynamic architecture of hox gene clusters. Science 2011, 334, 222–225.
- Bowman, S.K.; Deaton, A.M.; Domingues, H.; Wang, P.I.; Sadreyev, R.I.; Kingston, R.E.; Bender, W. H3k27 modifications define segmental regulatory domains in the drosophila bithorax complex. eLife 2014, 3, e02833.
- Akam, M.; Dawson, I.; Tear, G. Homeotic genes and the control of segment diversity. Dev. Suppl. 1988, 104, 123–133.
- Maeda, R.K.; Karch, F. The open for business model of the bithorax complex in drosophila. Chromosoma 2015, 124, 293–307.
- Gehring, W.J.; Kloter, U.; Suga, H. Evolution of the hox gene complex from an evolutionary ground state. Curr. Top. Dev. Biol. 2009, 88, 35–61.
- Lewis, E.B. The bithorax complex: The first fifty years. Int. J. Dev. Biol. 1998, 42, 403–415.
- Tarailo-Graovac, M.; Chen, N. Gene Clustering in Eukaryotes. In Els; John Wiley Sons Ltd.: Chichester, UK, 2013.
- Gaunt, S.J.; Gaunt, A.L. Possible rules for the ancestral origin of hox gene collinearity. J. Theor. Biol. 2016, 410, 1–8.
- Holland, P.W.H. Evolution of homeobox genes. Wires Dev. Biol. 2013, 2, 31–45.
- Hui, J.H.L.; McDougall, C.; Monteiro, A.S.; Holland, P.W.H.; Arendt, D.; Balavoine, G.; Ferrier, D.E.K. Extensive chordate and annelid macrosynteny reveals ancestral homeobox gene organization. Mol. Biol. Evol. 2012, 29, 157–165.
- Pollard, S.L.; Holland, P.W.H. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr. Biol. 2000, 10, 1059–1062.
- Garcia-Fernandez, J. The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 2005, 6, 881–892.
- Ferrier, D.E. The origin of the hox/parahox genes, the ghost locus hypothesis and the complexity of the first animal. Brief. Funct. Genom. 2016, 15, 333–341.
- Kamm, K.; Schierwater, B.; Jakob, W.; Dellaporta, S.L.; Miller, D.J.; Axial patterning and diversification in the cnidaria predate the hox system. current biology 2006, 16, 920-926.
- Derelle, R.; Lopez, P.; Le Guyader, H.; Manuel, M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol. Dev. 2007, 9, 212–219.
- Negre, B.; Ruiz, A. Hom-c evolution in drosophila: Is there a need for hox gene clustering? Trends Genet. TIG 2007, 23, 55–59.
- Shippy, T.D.; Hosmani, P.S.; Florez-Gonzalez, M.; Mueller, L.A.; Hunter, W.B.; Brown, S.J.; D’Elia, T.; Saha, S. Annotation of hox cluster and hox cofactor genes in the asian citrus psyllid, diaphorina citri, reveals novel features. Gigabyte 2022.
- Seo, H.C.; Edvardsen, R.B.; Maeland, A.D.; Bjordal, M.; Jensen, M.F.; Hansen, A.; Flaat, M.; Weissenbach, J.; Lehrach, H.; Wincker, P.; et al. Hox cluster disintegration with persistent anteroposterior order of expression in oikopleura dioica. Nature 2004, 431, 67–71.
- Celniker, S.E.; Sharma, S.; Keelan, D.J.; Lewis, E.B. The molecular genetics of the bithorax complex of drosophila: Cis-regulation in the abdominal-b domain. EMBO J. 1990, 9, 4277–4286.
- Gould, A.; Morrison, A.; Sproat, G.; White, R.A.; Krumlauf, R. Positive cross-regulation and enhancer sharing: Two mechanisms for specifying overlapping hox expression patterns. Genes Dev. 1997, 11, 900–913.
- Kwan, C.T.; Tsang, S.L.; Krumlauf, R.; Sham, M.H. Regulatory analysis of the mouse hoxb3 gene: Multiple elements work in concert to direct temporal and spatial patterns of expression. Dev. Biol. 2001, 232, 176–190.
- Montavon, T.; Duboule, D. Chromatin organization and global regulation of hox gene clusters. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120367.
- Duboule, D. The rise and fall of hox gene clusters. Development 2007, 134, 2549–2560.
- Pace, R.M.; Grbic, M.; Nagy, L.M. Composition and genomic organization of arthropod hox clusters. EvoDevo 2016, 7, 11.
- Sun, D.A.; Patel, N.H. The amphipod crustacean parhyale hawaiensis: An emerging comparative model of arthropod development, evolution, and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2019, 8, e355.
- Dearden, P.K.; Wilson, M.J.; Sablan, L.; Osborne, P.W.; Havler, M.; McNaughton, E.; Kimura, K.; Milshina, N.V.; Hasselmann, M.; Gempe, T.; et al. Patterns of conservation and change in honey bee developmental genes. Genome Res. 2006, 16, 1376–1384.
- Shippy, T.D.; Ronshaugen, M.; Cande, J.; He, J.; Beeman, R.W.; Levine, M.; Brown, S.J.; Denell, R.E. Analysis of the tribolium homeotic complex: Insights into mechanisms constraining insect hox clusters. Dev. Genes Evol. 2008, 218, 127–139.
- Freeman, R.; Ikuta, T.; Wu, M.; Koyanagi, R.; Kawashima, T.; Tagawa, K.; Humphreys, T.; Fang, G.C.; Fujiyama, A.; Saiga, H.; et al. Identical genomic organization of two hemichordate hox clusters. Curr. Biol. CB 2012, 22, 2053–2058.
- Arenas-Mena, C.; Cameron, A.R.; Davidson, E.H. Spatial expression of hox cluster genes in the ontogeny of a sea urchin. Development 2000, 127, 4631–4643.
- Jorgensen, E.M.; Garber, R.L. Function and misfunction of the two promoters of the drosophila antennapedia gene. Genes Dev. 1987, 1, 544–555.
- Schneuwly, S.; Kuroiwa, A.; Gehring, W.J. Molecular analysis of the dominant homeotic antennapedia phenotype. EMBO J. 1987, 6, 201–206.
- Darbellay, F.; Bochaton, C.; Lopez-Delisle, L.; Mascrez, B.; Tschopp, P.; Delpretti, S.; Zakany, J.; Duboule, D. The constrained architecture of mammalian hox gene clusters. Proc. Natl. Acad. Sci. USA 2019, 116, 13424–13433.
- Izpisua-Belmonte, J.C.; Falkenstein, H.; Dolle, P.; Rennuci, A.; Duboule, D.; Murine genes related to the drosophila abdb homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991, 10, 2279-2289.
- Pascual-Anaya, J.; Adachi, N.; Alvarez, S.; Kuratani, S.; D’Aniello, S.; Garcia-Fennandez, J.; Broken colinearity of the amphioxus hox cluster. EvoDevo 2012, 3, 28.
- Frobius, A.C.; Matus, D.Q.; Seaver, E.C.; Genomic organization and expression demonstrate spatial and temporal hox gene colinearity in the lophotrochozoan capitella sp. I . Plos one 2008, 3, e4004.
- Serano, J.M.; Martin, A.; Liubicich, D.M.; Jarvis, E.; Bruce, H.S.; La, K.; Grimwood, J.; Patel, N.H.; Comprehensive analysis of hox gene expression in the amphipod crustacean parhyale hawaiensis.. Dev. Biol. 2016, 409, 297-309.
- Duboule, D. Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994, 1994, 135–142.
- Ferrier, D.E.; Holland, P.W. Ciona intestinalis parahox genes: Evolution of hox/parahox cluster integrity, developmental mode, and temporal colinearity. Mol. Phylogenet. Evol. 2002, 24, 412–417.
- Gold, D.A.; Runnegar, B.; Gehling, J.G.; Jacobs, D.K. Ancestral state reconstruction of ontogeny supports a bilaterian affinity for dickinsonia. Evol. Dev. 2015, 17, 315–324.
- Rekaik, H.; Lopez-Delisle, L.; Hintermann, A.; Mascrez, B.; Bochaton, C.; Duboule, D. Sequential and directional insulation by conserved ctcf sites underlies the hox timer in pseudo-embryos. bioRkiv 2022.
- Amandio, A.R.; Beccari, L.; Lopez-Delisle, L.; Mascrez, B.; Zakany, J.; Gitto, S.; Duboule, D. Sequential in cis mutagenesis in vivo reveals various functions for ctcf sites at the mouse hoxd cluster. Genes Dev. 2021, 35, 1490–1509.
- Duboule, D. The (unusual) heuristic value of hox gene clusters; a matter of time? Dev. Biol. 2022, 484, 75–87.
- Wei, M.; Qin, Z.; Kong, D.; Liu, D.; Zheng, Q.S.; Bai, S.; Zhang, Z.; Ma, Y. Echiuran hox genes provide new insights into the correspondence between hox subcluster organization and collinearity pattern. Proc. R. Soc. B 2022, 289, 20220705.
- Aronowicz, J.; Lowe, C.J.; Hox gene expression in the hemichordate saccoglossus kowalevskii and the evolution of deuterostome nervous systems. Integrative and comparative biology 2006, 46, 890-901.




