| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yu-Fan Chen | + 921 word(s) | 921 | 2020-11-18 09:52:12 | | | |
| 2 | Yu-Fan Chen | + 2 word(s) | 923 | 2020-12-17 12:45:59 | | | | |
| 3 | Vicky Zhou | -1 word(s) | 922 | 2020-12-18 03:23:08 | | |
Video Upload Options
The microbial cell–cell chemical communication systems include QS and host–pathogen communication systems. In 1994, Fuqua et al., coined the term of “quorum sensing” to describe an environmental sensing system that monitors the population density to coordinate the social behaviors of microorganisms. QS regulatory systems are characterized by the fact that microorganisms produce and release to the surrounding environment a diffffusible autoinducer or QS signal, which accumulates along with bacterial growth and induces target gene transcriptional expression when reaching a threshold concentration. Typically, a QS system contains a signal synthase, which produces QS signals, and a signal receptor that detect and response to QS signal in a population-dependent manner. In addition, microbial pathogens can also exploit the chemical molecules produced by host organisms as cross-kingdom signals to regulate virulence gene expression.
1. AHL-Type QS System
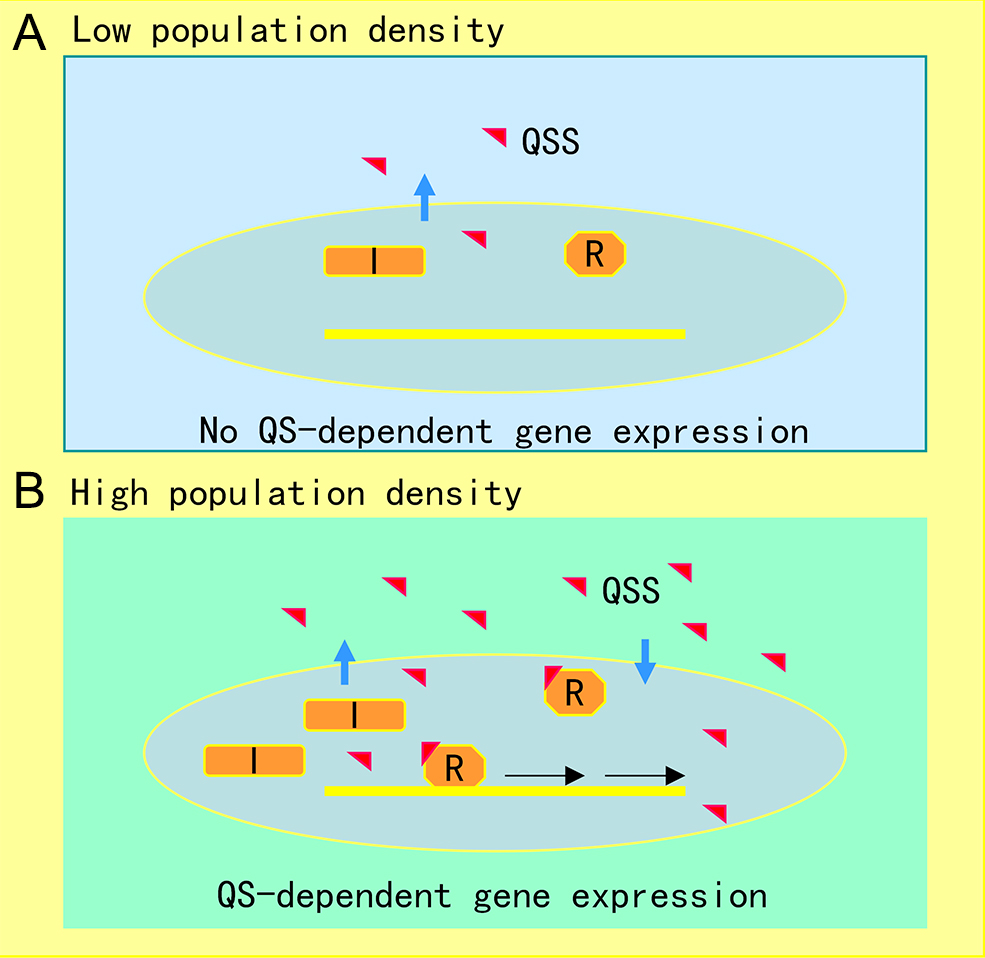
The AHL-mediated QS system is one of the most studied cell-to-cell communication systems, with over 200 bacterial species known to produce AHL family signals. Different bacterial species produce similar AHL signaling molecules with a conserved N-acyl homoserine lactone, but the fatty acid side chain lengths and substituents are in general variable, which may account for their signaling specificity in different microorganisms [1]. A wide range of microbial biological functions are known to be regulated by the AHL QS system, including bioluminescence, plasmid DNA transfer, production of pathogenic factors, biofilm formation, and antibiotic production [2]. The core of the AHL QS system consists of the AHL synthase (LuxI homologue) and the AHL signal receptor (LuxR homologue). In the bioluminescent bacterium A. fischeri, where the AHL QS system was first identified, at low cell density, the LuxI protein produces a basal level of few diffusible AHL molecules, and little bioluminescence could be detected. In contrast, with an increase in bacterial population density, the AHL signal molecules that accumulated around the bacterial cells enter the cell and bind and activate the LuxR protein, which then initiates LuxI overexpression and activates transcription of luminescence genes [3]. The generic mechanism of AHL-mediated QS is illustrated in Figure 1.
Figure 1. Illustration of quorum sensing (QS) mechanism in microorganisms. (A) At low cell population density, bacterial cells produce limited QS signal (QSS) which cannot trigger QS-dependent gene expression. (B) Along with bacterial growth, accumulated QSS interacts with and hence activates its cognate receptor to induce virulence factor production, biofilm formation, and generation of efflux pumps, which aid the pathogen survival in the host by counteracting various possible stresses, including immune responses and antibiotics. Symbols: I represents the QS signal synthase, R is the receptor, and the red triangle indicates QS signal.
2. DSF-type QS system
DSF represents another class of QS systems that are widespread in Gram-negative bacteria. In 1997, the Daniels Laboratory in the UK discovered that plant pathogen Xanthomonas campestris pv.campesitris (Xcc) could produce a diffusible signaling factor (DSF) and regulate production of a number of virulence factors, including proteases, cellulases and extracellular polysaccharides [4][5]. A subsequent study showed that three genes are associated with the DSF QS system, in which the rpfF gene may encode a DSF synthase, and the products of rpfC and rpfG genes could be involved in DSF signaling [5]. In 2004, Wang et al. isolated and identified DSF as cis-11-methyl-2-dodecenoic acid [6]. To date, more than 10 structurally similar DSF family signals have been isolated and identified from Gram-negative bacteria, and over 100 bacterial species were predicted to use DSF to regulate various biological functions [7][8]. The biological functions and signaling mechanisms of the DSF systems have been well studied in Xcc and Burkholderia cenocepacia [9][10]. In Xcc, response to DSF signals leads to activation of the protein kinase RpfC by self-phosphorylation, which activates the phosphodiesterase activity of its cognate response regulator RpfG through phosphorelay; the activated RpfG degrades c-di-GMP molecules and activates the global regulator Clp; Clp directly regulates the transcription of a number of pathogenic genes and indirectly regulates the expression of other pathogenic genes through the downstream transcription factors FhrR and Zur [9]. In B. cenocepacia, another DSF family signal, cis-2-dodecenoic acid (BDSF), binds to its major receptor RpfR to reduce the intracellular c-di-GMP to a sufficient level, which stimulate the RpfR-GtrR protein complex to bind to the promoter region of target DNA, thus modulating the production of virulence factors and biofilm mass [11][12][13].
3. Polyamine-Mediated Host–Pathogen Communication Systems
Spermidine (Spd), spermine (Spm), and putrescine (Put) represent a widespread group of cationic aliphatic compounds with multiple biological activities in living cells, collectively referred to as polyamines. In recent years, increasing evidence indicates that polyamines are capable of performing regulatory functions as signals in intraspecies cell–cell communication or are involved in host–pathogen trans-kingdom cell-cell communication [14][15]. The precursors for synthesis of Put, Spd and Spm in bacterial cells are l-arginine, l-ornithine and l-methionine, respectively [16]. l-arginine decarboxylase decarboxylates arginine to produce adamantane, which is then converted to Put and urea by agmatine ureohydrolase. l-ornithine is decarboxylated by ornithine decarboxylase (ODC) to produce Put, then Put undergoes a transfer reaction with a propylamine group by the action of arginine synthetase to produce Spd, and then undergoes another transfer reaction with a propylamine group by the action of Spm synthetase to produce Spm [17]. l-methionine is catalyzed by l-methionine adenosyltransferase to produce S-adenosylmethionine (SAM), which is then decarboxylated by S-adenosylmethionine decarboxylase (SAMDC) to produce S-adenosylmethionine (dcSAM), which provides the propylamine group for the synthesis of Spd and Spm [17]. In bacteria, polyamines are mainly involved in the regulation of important physiological processes such as transcription and translation of bacterial virulence factors, biofilm formation, antibiotic resistance, acidic and oxidative stresses [16][18].
References
- Blevins, S.M.; Bronze, M.S. Robert Koch and the ‘golden age’ of bacteriology. Int. J. Infect. Dis. 2010, 14, e744–e751.
- Berche, P. Louis Pasteur, from crystals of life to vaccination. Clin. Microbiol. Infect. 2012, 18 (Suppl. 5), 1–6.
- Schwartz, R.S. Paul Ehrlich’s magic bullets. N. Engl. J. Med. 2004, 350, 1079–1080.
- Hugh, T.B. Howard Florey, Alexander Fleming and the fairy tale of penicillin. Med. J. Aust. 2002, 177, 52–53.
- Mohr, K.I. History of Antibiotics Research. Curr. Top. Microbiol. Immunol. 2016, 398, 237–272.
- Waglechner, N.; McArthur, A.G.; Wright, G.D. Phylogenetic reconciliation reveals the natural history of glycopeptide antibiotic biosynthesis and resistance. Nat. Microbiol. 2019, 4, 1862–1871.
- Gould, K. Antibiotics: From prehistory to the present day. J. Antimicrob. Chemother. 2016, 71, 572–575.
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281.
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910.
- Spellberg, B.; Guidos, R.; Gilbert, D.; Bradley, J.; Boucher, H.W.; Scheld, W.M.; Bartlett, J.G.; Edwards, J., Jr.; Infectious Diseases Society of, A. The epidemic of antibiotic-resistant infections: A call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 155–164.
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461.
- von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173.
- Dickey, S.W.; Cheung, G.Y.C.; Otto, M. Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017, 16, 457–471.
- Anes, J.; McCusker, M.P.; Fanning, S.; Martins, M. The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 2015, 6, 587.
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267.
- Deng, Z.; Shan, Y.; Pan, Q.; Gao, X.; Yan, A. Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front. Microbiol. 2013, 4, 194.
- Borlee, B.R.; Goldman, A.D.; Murakami, K.; Samudrala, R.; Wozniak, D.J.; Parsek, M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010, 75, 827–842.
- Chambers, J.R.; Sauer, K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013, 21, 39–49.