
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Renpeng Du | -- | 3068 | 2022-11-24 09:18:30 | | | |
| 2 | Camila Xu | + 2 word(s) | 3070 | 2022-11-25 03:57:15 | | |
Video Upload Options
Glucansucrase (GS) belongs to the GH70 family, which not only can synthesize exopolysaccharides (EPSs) with different physicochemical properties through glucosyl transglycosylation (by hydrolyzing sucrose) but can also produce oligosaccharides.
1. Introduction
2. Structure of the GS
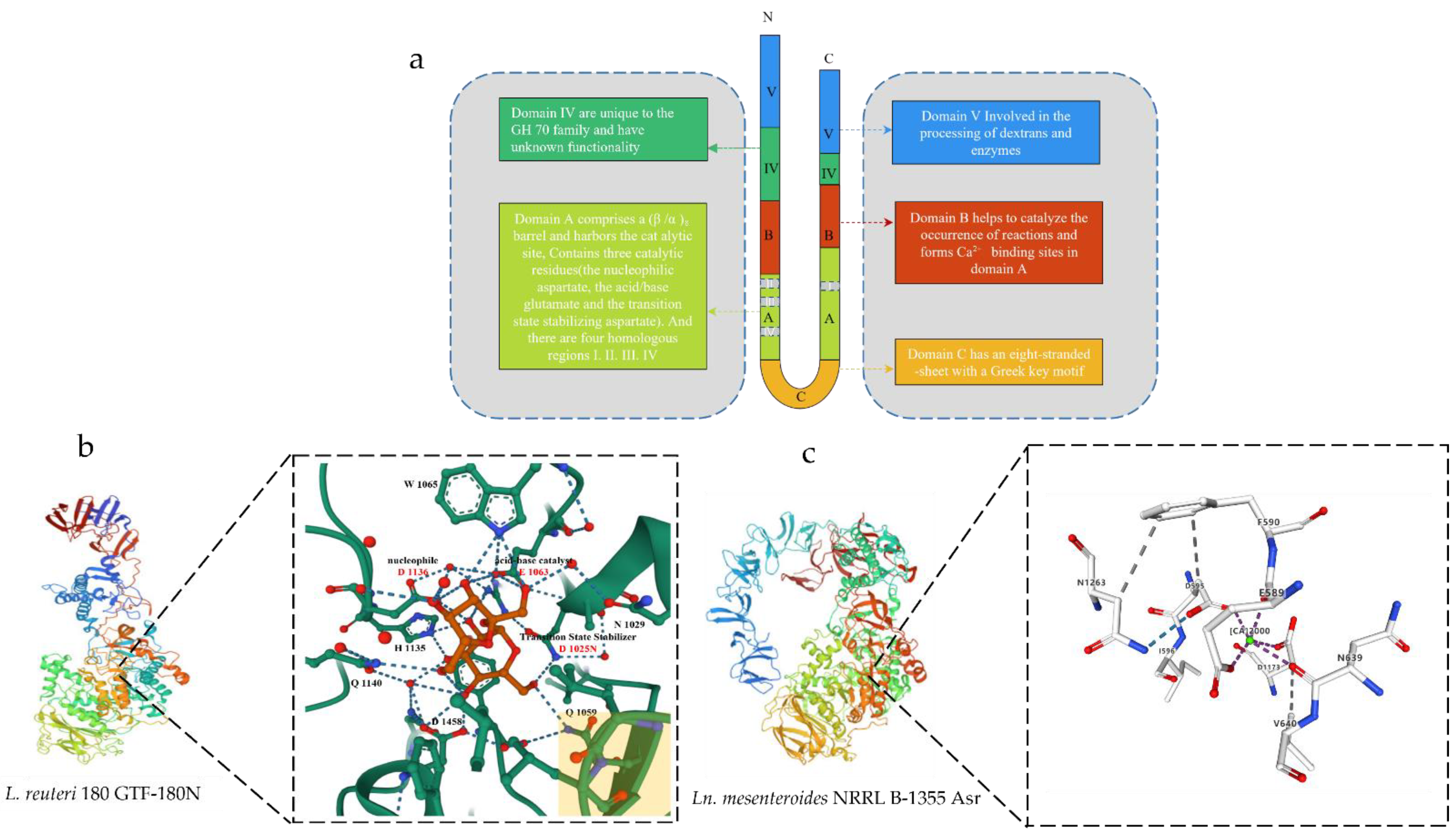
Figure 1. Schematic diagram of the 3D structure of GS. (a) Schematic representation of the U-shape fold formed by the various domains of GS. (b) Schematic diagram of the 3D structure of Ln. reuteri GTF-180N (PDB: 3HZ3) and sucrose binding site. Sucrose molecules are shown in red, residues in domain A are shown in green carbon, and residues in domain B are shown in yellow at the bottom right. (c) Schematic diagram of the 3D structure of Ln. mesenteroides NRRL B-1355 Asr (PDB: 6HVG) and Ca2+ binding sites. The green circles represent Ca2+.
3. Catalytic Mechanism of GS
Both the catalytic mechanism and structure of GH70 are closely related to the GH13 and GH77 families [24]. The newly discovered GH70 subfamily GtfB, GtfC, and GtfD are inactive against sucrose but can catalyze starch and maltodextrin to α-glucan [24]. The common characteristic of GH family enzymes is that they also use a catalytic (β/α)8 barrel domain to break down the α-glycosidic bonds between glucose and other glucose or fructose [29][30]. Robyt found there are two active sites; according to the GS catalytic reaction, one is composed of covalent β-glucose-enzyme intermediates under oxygen-carbon ion-like transformation conditions [5][31]. The β-glucose-enzyme intermediate is catalyzed through the a-retaining double displacement reaction, which involves three important residues: a nucleophile, an acid-base catalyst, and a transition state stabilizer (Figure 2). Aspartic acid, which first acts as a nucleophile, attacks the ectopic C1 carbon of the sucrose–glucose unit. Glutamate acts as an acid-base catalyst to transfer protons to fructose and release fructose. The transition state stabilizes the stable residue dimension and transitions to a covalent β-glucose-enzyme. Finally, the covalent β-glucosyl-enzyme is formed from the transition state stabilizer (Figure 2). The other is composed of chain and enzyme intermediates [30]. The C1 position in the later intermediate attacks the C6 position of the glucosyl group to form a glycosidic bond, thereby increasing the length [31]. Although the determinants of the size distribution of GS products have been broadly studied before, many are still unknown [26][32][33][34][35][36]. The N-terminal variable region and C-terminal glucan-binding domain have been indicated as playing a role in product size distribution [26].
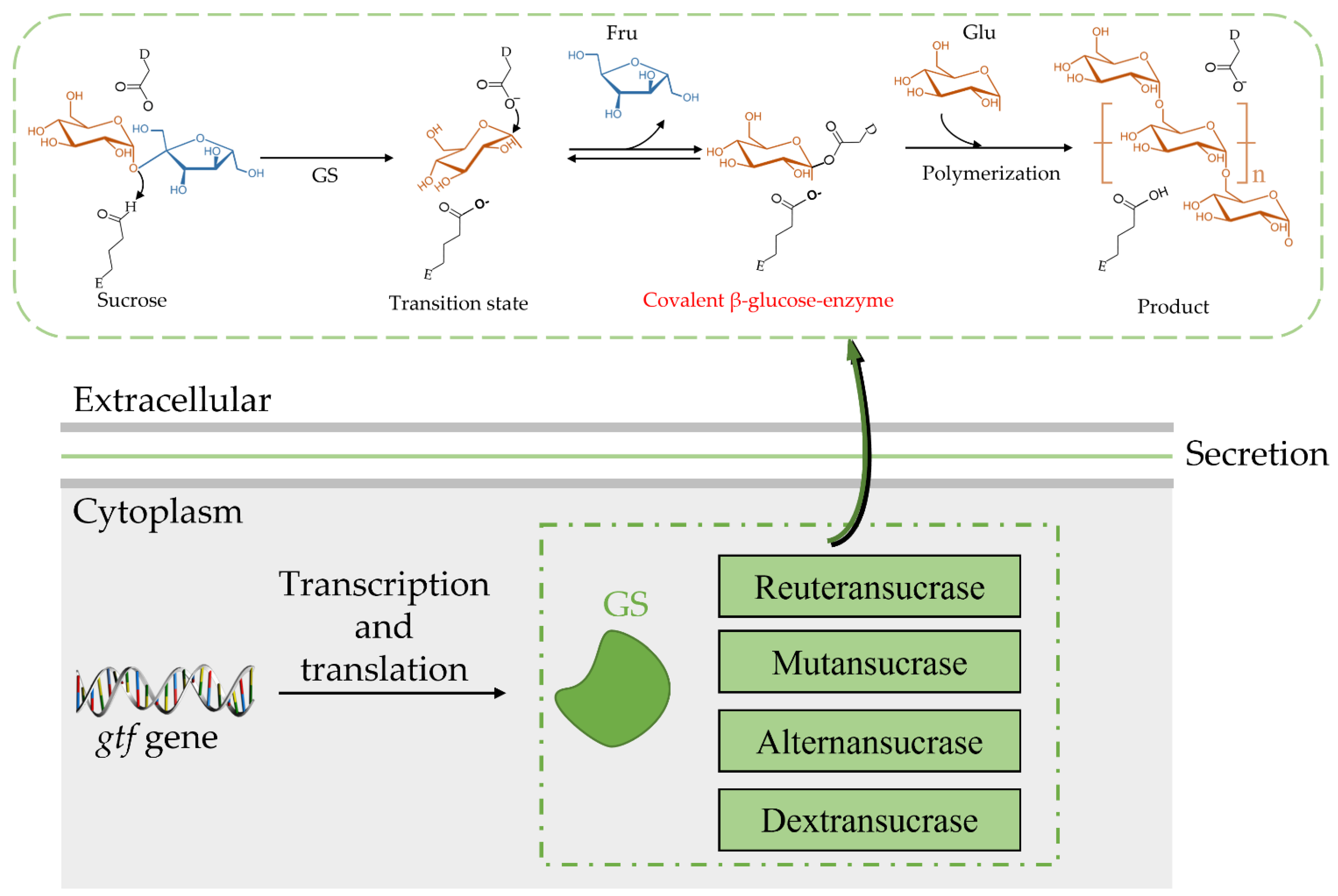
4. Isolation and Purification of GS
| Enzyme | MASS | Organism | Genebank | Length | Reference |
|---|---|---|---|---|---|
| Gtf1624 | 183 kDa | Latilactobacillus curvatus TMW 1.624 | CCK33643.1 | 1697 aa | [57] |
| DSR-F | 170 kDa | Ln. citreum B/110-1-2 | ACY92456.2 | 1527 aa | [58] |
| LcDS | 165 kDa | Ln. citreum HJ-P4 | BAF96719.1 | 1477 aa | [59] |
| DexT | 167 kDa | Ln citreum KM20 | ACA83218.1 | 1495 aa | [60] |
| DSR-A | 145 kDa | Ln. citreum NRRL B-1299 | CDX67012.1 | 1290 aa | [61] |
| DSR-B | 168 kDa | Ln. citreum NRRL B-1299 | AAB95453.1 | 1508 aa | [62] |
| DSR-E | 313 kDa | Ln. citreum NRRL B-1299 | CDX66820.1 | 2836 aa | [63] |
| DSR-DP | - | Ln. citreum NRRL B-1299 | CDX66641.1 | 1278 aa | [63] |
| DSR-M | 144 kDa | Ln. citreum NRRL B-1299 | CDX66895.1 | 1293 aa | [61] |
| DexYG | 170 kDa | Ln. mesenteroides 0326 | ABC75033.1 | 1527 aa | [64] |
| DsrBCB4 | 168 kDa | Ln. mesenteroides B-1299CB4 | ABF85832.1 | 1505 aa | [65] |
| DsrC | 165 kDa | Ln. mesenteroides B-1355 | CAB76565.1 | 1477 aa | [66] |
| Dsrb74 | 169 kDa | Ln. mesenteroides B-742CB | AAG38021.1 | 1508 aa | [67] |
| DsrP | 161 kDa | Ln. mesenteroides IBT-PQ | AAS79426.1 | 1454 aa | [68] |
| DsrN | 169 kDa | Ln. mesenteroides KIBGE-IB-22 | AFP53921.1 | 1527 aa | [69] |
| DsrX | 169 kDa | Ln. mesenteroides L0309 | AAQ98615.2 | 1522 aa | [70] |
| DsrD | 169 kDa | Ln. mesenteroides LCC4 | AAG61158.1 | 1527 aa | [50] |
| DSR-S | 169 kDa | Ln. mesenteroides NRRL B-512F | AAD10952.1 | 1527 aa | [71] |
| DSR-T | 110 kDa | Ln. mesenteroides NRRL B-512F | BAA90527.1 | 1016 aa | [72] |
| Gtf1971 | 178 kDa | Ligilactobacillus animalis TMW 1.971 | CCK33644.1 | 1585 aa | [57] |
| Gtf106A | 199 kDa | L. reuteri TMW 1.106 | ABP88726.1 | 1782 aa | [57] |
| GTF-S | 151 kDa | Streptococcus downei MFE 28 | AAA26898.1 | 1365 aa | [73] |
| GTF-U | 176 kDa | Streptococcus sobrinus | BAA14241.1 | 1592 aa | [74] |
| GTF-B | 166 kDa | Streptococcus mutans GS 5 | AAA88588.1 | 1476 aa | [75] |
| GTF-D | 163 kDa | S. mutans GS 5 | AAA26895.1 | 1462 aa | [76] |
| DsrK39 | 158 kDa | Weissella cibaria LBAE-K39 | ADB43097.3 | 1445 aa | [77] |
| WcCab3-DSR | 154 kDa | Weissella confusa Cab3 | AKE50934.1 | 1401 aa | [78] |
| DSR-C39-2 | 155 kDa | W. confusa LBAE C39-2 | CCF30682.1 | 1412 aa | [54] |
| DSR | 156 kDa | W. confusa VTT E-90392 | AHU88292.1 | 1418 aa | [79] |
| LcALT | 229 kDa | Ln. citreum ABK-1 | AIM52834.1 | 2057 aa | [80] |
| GtfB-SK2 | - | Ln. citreum SK24.002 | - | - | [81] |
| ASR | 229 kDa | Ln. mesenteroides NRRL B-1355 | CAB65910.2 | 2057 aa | [82] |
| GtfO | 197 kDa | L. reuteri ATCC 55730 | AAY86923.1 | 1781 aa | [83] |
| Gtf-SK3 | - | L. reuteri SK24.003 | - | - | [84] |
| GtfML1 | - | L. reuteri ML1 | - | - | [34] |
| DSRI | - | Ln. mesenteroides NRRL B-1118 | - | - | [85] |
| GtfB | - | S. mutans GS5 | - | - | [86] |
| GtfC | - | S. mutans GS5 | - | - | [87] |
| GTF-Kg15 | 174 kDa | Latilactobacillus sakei KG15 | AAU08011.1 | 1595 aa | [34] |
| GTF-33 | 172 kDa | Lentilactobacillus parabuchneri 33 | AAU08006.1 | 1561 aa | [34] |
| - | 164 kDa | Leuconostoc lactis EG001 | ACT20911.1 | 1500 aa | [56] |
| GTF-Kg3 | 161 kDa | Limosilactobacillus fermentum KG3 | AAU08008.1 | 1463 aa | [34] |
| GtfB | 179 kDa | L. f ermentum NCC2970 | AOR73699.1 | 1593 aa | [88] |
| GtfB | 179 kDa | L. reuteri 121 | AAU08014.2 | 1619 aa | [89] |
| GtfML1 | 196 kDa | L. reuteri ML1 | AAU08004.1 | 1772 aa | [34] |
| GtfML4 | 180 kDa | L. reuteri ML1 | AAU08003.2 | 1620 aa | [90] |
| GtfB | 196 kDa | L. reuteri NCC2613 | ASA47879.1 | 1662 aa | [91] |
| GtfC | 163 kDa | S. mutans | BAA26114.1 | 1455 aa | [89] |
| DsrwC | 162 kDa | W. cibaria CMU | ACK38203.1 | 1472 aa | [92] |
| GtfD | 87 kDa | Azotobacter chroococcum NCIMB 8003 | AJE22990.1 | 780 aa | [93] |
| GtfC | 99 kDa | Exiguobacterium sibiricum 255-15 | ACB62096.1 | 893 aa | [94] |
5. Physiological and Biochemical Properties of GS
Many factors can affect the catalytic activity of GS, including pH, temperature, and some organic solvents and metal ions. According to Kralj [34], the maximum GS activity from L. reuteri was gained at a pH of 4.0–5.5, which is comparable with most papers. Miao [52] found that GS from L. reuteri SK24.003 retained high activity at a low pH, showing better acid-resistance. However, a pH value that is too high or too low is still not conducive to the synthesis of GS. Previous studies have shown that GS has the highest purity and the most stable activity at pH 7 [95], while a low pH can significantly reduce enzyme synthesis. The analysis of the GS produced by Ln. mesenteroides DRP2-19 and isolated from sauerkraut showed that the optimum pH for GS was 5.56, and the synthesis of GS was severely affected at pH < 4.5 or >7, which corroborated the previous point [96]. Because of the different sources and structures of GSs, the optimum reaction temperature for each GS is different. Generally, the optimum range is from 30–40°C in culture. When the temperature exceeds 45 °C, the enzyme activity begins to decrease, and when it exceeds 50 °C, the enzyme loss is more obvious. Most double-charged ions (Mg2+, Mn2+, Ni2+, Co2+, Ca2+, Fe2+, and Zn2+) activated enzyme activity, suggesting it was a metal-activated enzyme [52]. GS has a calcium ion activation site, which can increase enzyme activity. It is concluded that calcium ion can enhance the activity of dextransucrase. The stability of the entire edifice is enhanced by calcium coordination, which likely reinforces the interaction between the two domains [97]. Qader et al. [98] found that when the concentration of CaCl2 was 0.005%, the enzyme activity increased to 108.26 DSU/mL/h, which was 2.03 times higher than that of the control group. When the content of CaCl2 was higher than 0.005%, enzyme activity decreased gradually. Some reports found that when Ca2+ is within a certain concentration range, Ca2+ will preferentially bind to the activation site on the enzyme, and the activation effect is stronger than the inhibitory effect. Furthermore, Hg+, Zn2+, Cu2+, Pb2+, and Fe3+ had a strong inhibitory effect on enzyme activity, which was the same as in previous reports; when copper ions were present, there was no transferase activity [99]. The activity and stability of GS in the presence of organic solvents were related to the solvent concentration and its nature. Chemical inhibitors indicate that the function of amino acid residues is located at the GS-active site [16]. Most chemical inhibitors had an inhibition effect on GS, like sodium dodecyl sulfate (SDS), ethylene diamine, tetraacetic acid (EDTA), and β-Mercaptoethanol (β-ME) [25][100]. Other chemical reagents, including butanol, n-hexane, chloroform, calcium ammonium nitrate (CAN), and ethyl acetate, also inhibited the activity of GS with increasing concentrations, whereas glycerol, formaldehyde, and dimethyl sulfoxide (DMSO) enhanced the activity of GS to a certain extent [101].
References
- Xu, Y.; Cui, Y.; Yue, F.; Liu, L.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Exopolysaccharides produced by lactic acid bacteria and Bifidobacteria: Structures, physiochemical functions and applications in the food industry. Food Hydrocoll. 2019, 94, 475–499.
- Rana, S.; Upadhyay, L.S.B. Microbial exopolysaccharides: Synthesis pathways, types and their commercial applications. Int. J. Biol. Macromol. 2020, 157, 577–583.
- Pham, H.; Pijning, T.; Dijkhuizen, L.; van Leeuwen, S.S. Mutational analysis of the role of the glucansucrase Gtf180-ΔN active site residues in product and linkage specificity with lactose as acceptor substrate. J. Agric. Food Chem. 2018, 66, 12544–12554.
- Meng, X.; Pijning, T.; Tietema, M.; Dobruchowska, J.M.; Yin, H.; Gerwig, G.J.; Kralj, S.; Dijkhuizen, L. Characterization of the glucansucrase GTF180 W1065 mutant enzymes producing polysaccharides and oligosaccharides with altered linkage composition. Food Chem. 2017, 217, 81–90.
- Molina, M.; Cioci, G.; Moulis, C.; Séverac, E.; Remaud-Siméon, M. Bacterial α-glucan and branching sucrases from GH70 family: Discovery, structure–function relationship studies and engineering. Microorganisms 2021, 9, 1607.
- Holt, S.M.; Skory, C.; Cote, G. Enzymatic synthesis of artificial polysaccharides. ACS Sustain. Chem. Eng. 2020, 8, 11853–11871.
- Chen, Z.; Ni, D.; Zhang, W.; Stressler, T.; Mu, W. Lactic acid bacteria-derived α-glucans: From enzymatic synthesis to miscellaneous applications. Biotechnol. Adv. 2021, 47, 107708.
- Kim, M.; Jang, J.-K.; Park, Y.-S. Production optimization, structural analysis, and prebiotic-and anti-inflammatory effects of gluco-oligosaccharides produced by Leuconostoc lactis SBC001. Microorganisms 2021, 9, 200.
- Hasselwander, O.; DiCosimo, R.; You, Z.; Cheng, Q.; Rothman, S.C.; Suwannakham, S.; Baer, Z.C.; Roesch, B.M.; Ruebling-Jass, K.D.; Lai, J.P. Development of dietary soluble fibres by enzymatic synthesis and assessment of their digestibility in vitro, animal and randomised clinical trial models. Int. J. Food Sci. Nutr. 2017, 68, 849–864.
- Saadat, Y.R.; Khosroushahi, A.Y.; Gargari, B.P. A comprehensive review of anticancer, immunomodulatory and health beneficial effects of the lactic acid bacteria exopolysaccharides. Carbohydr. Polym. 2019, 217, 79–89.
- Sarbini, S.R.; Kolida, S.; Naeye, T.; Einerhand, A.W.; Gibson, G.R.; Rastall, R.A. The prebiotic effect of α-1, 2 branched, low molecular weight dextran in the batch and continuous faecal fermentation system. J. Funct. Foods 2013, 5, 1938–1946.
- Sarbini, S.R.; Kolida, S.; Naeye, T.; Einerhand, A.; Brison, Y.; Remaud-Simeon, M.; Monsan, P.; Gibson, G.R.; Rastall, R.A. In vitro fermentation of linear and α-1, 2-branched dextrans by the human fecal microbiota. Appl. Environ. Microbiol. 2011, 77, 5307–5315.
- Daba, G.M.; Elnahas, M.O.; Elkhateeb, W.A. Contributions of exopolysaccharides from lactic acid bacteria as biotechnological tools in food, pharmaceutical, and medical applications. Int. J. Biol. Macromol. 2021, 173, 79–89.
- Korcz, E.; Kerényi, Z.; Varga, L. Dietary fibers, prebiotics, and exopolysaccharides produced by lactic acid bacteria: Potential health benefits with special regard to cholesterol-lowering effects. Food Funct. 2018, 9, 3057–3068.
- Kabli, M.; İspirli, H.; Balubaid, M.; Taylan, O.; Yılmaz, M.T.; Dertli, E. Optimization of lactose derivative hetero-oligosaccharides production using whey as the acceptor molecule by an active glucansucrase. Biocatal. Biotransform. 2022, 40, 9–16.
- Song, L.; Miao, M.; Jiang, B.; Xu, T.; Cui, S.W.; Zhang, T. Leuconostoc citreum SK24.002 glucansucrase: Biochemical characterisation and de novo synthesis of α-glucan. Int. J. Biol. Macromol. 2016, 91, 123–131.
- Guzman, G.Y.F.; Hurtado, G.B.; Ospina, S.A. New dextransucrase purification process of the enzyme produced by Leuconostoc mesenteroides IBUN 91.2. 98 based on binding product and dextranase hydrolysis. J. Biotechnol. 2018, 265, 8–14.
- Vasileva, T.; Bivolarski, V.; Michailova, G.; Salim, A.; Rabadjiev, Y.; Ivanova, I.; Iliev, I. Glucansucrases produced by fructophilic lactic acid bacteria Lactobacillus kunkeei H3 and H25 isolated from honeybees. J. Basic Microbiol. 2017, 57, 68–77.
- Meng, X.; Li, X.; Pijning, T.; Wang, X.; van Leeuwen, S.S.; Dijkhuizen, L.; Chen, G.; Liu, W. Characterization of the (engineered) branching sucrase gtfz-cd2 from Apilactobacillus kunkeei for efficient glucosylation of benzenediol compounds. Appl. Environ. Microbiol. 2022, 88, e01031-22.
- Vujičić-Žagar, A.; Pijning, T.; Kralj, S.; López, C.A.; Eeuwema, W.; Dijkhuizen, L.; Dijkstra, B.W. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21406–21411.
- Ito, K.; Ito, S.; Shimamura, T.; Weyand, S.; Kawarasaki, Y.; Misaka, T.; Abe, K.; Kobayashi, T.; Cameron, A.D.; Iwata, S. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 2011, 408, 177–186.
- Stam, M.R.; Danchin, E.G.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of α-amylase-related proteins. Protein Eng. Des. Sel. 2006, 19, 555–562.
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. Biotechnological potential of novel glycoside hydrolase family 70 enzymes synthesizing α-glucans from starch and sucrose. Biotechnol. Adv. 2018, 36, 196–207.
- Meng, X.; Gangoiti, J.; Bai, Y.; Pijning, T.; Van Leeuwen, S.S.; Dijkhuizen, L. Structure–function relationships of family GH70 glucansucrase and 4, 6-α-glucanotransferase enzymes, and their evolutionary relationships with family GH13 enzymes. Cell. Mol. Life Sci. 2016, 73, 2681–2706.
- Wu, D.T.; Zhang, H.B.; Huang, L.J.; Hu, X.Q. Purification and characterization of extracellular dextranase from a novel producer, Hypocrea lixii F1002, and its use in oligodextran production. Process Biochem. 2011, 46, 1942–1950.
- Moulis, C.; Vaca Medina, G.; Suwannarangsee, S.; Monsan, P.; Remaud-Simeon, M.; Potocki-Veronese, G. One-step synthesis of isomalto-oligosaccharide syrups and dextrans of controlled size using engineered dextransucrase. Biocatal. Biotransform. 2008, 26, 141–151.
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272.
- van Hijum, S.A.; Kralj, S.; Ozimek, L.K.; Dijkhuizen, L.; van Geel-Schutten, I.G. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 157–176.
- André, I.; Potocki-Véronese, G.; Morel, S.; Monsan, P.; Remaud-Siméon, M. Sucrose-utilizing transglucosidases for biocatalysis. Carbohydr. Sustain. Dev. I 2010, 294, 25–48.
- Korakli, M.; Vogel, R.F. Structure/function relationship of homopolysaccharide producing glycansucrases and therapeutic potential of their synthesised glycans. Appl. Microbiol. Biotechnol. 2006, 71, 790–803.
- Robyt, J.F.; Yoon, S.H.; Mukerjea, R. Dextransucrase and the mechanism for dextran biosynthesis. Carbohydr. Res. 2008, 343, 3039–3048.
- Falconer, D.J.; Mukerjea, R.; Robyt, J.F. Biosynthesis of dextrans with different molecular weights by selecting the concentration of Leuconostoc mesenteroides B-512FMC dextransucrase, the sucrose concentration, and the temperature. Carbohydr. Res. 2011, 346, 280–284.
- Kim, D.; Robyt, J.F.; Lee, S.Y.; Lee, J.H.; Kim, Y.M. Dextran molecular size and degree of branching as a function of sucrose concentration, pH, and temperature of reaction of Leuconostoc mesenteroides B-512FMCM dextransucrase. Carbohydr. Res. 2003, 338, 1183–1189.
- Kralj, S.; van Geel-Schutten, G.H.; Dondorff, M.M.G.; Kirsanovs, S.; Van Der Maarel, M.; Dijkhuizen, L. Glucan synthesis in the genus Lactobacillus: Isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 2004, 150, 3681–3690.
- Lee, M.S.; Cho, S.K.; Eom, H.J.; Kim, S.Y.; Kim, T.J.; Han, N.S. Optimized substrate concentrations for production of long-chain isomaltooligosaccharides using dextransucrase of Leuconostoc mesenteroides B-512F. J. Microbiol. Biotechnol. 2008, 18, 1141–1145.
- Meng, X.; Dobruchowska, J.M.; Pijning, T.; Gerwig, G.J.; Kamerling, J.P.; Dijkhuizen, L. Truncation of domain V of the multidomain glucansucrase GTF180 of Lactobacillus reuteri 180 heavily impairs its polysaccharide-synthesizing ability. Appl. Microbiol. Biotechnol. 2015, 99, 5885–5894.
- Lawson, C.L.; van Montfort, R.; Strokopytov, B.; Rozeboom, H.J.; Kalk, K.H.; de Vries, G.E.; Penninga, D.; Dijkhuizen, L.; Dijkstra, B.W. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J. Mol. Biol. 1994, 236, 590–600.
- Kralj, S.; Eeuwema, W.; Eckhardt, T.H.; Dijkhuizen, L. Role of asparagine 1134 in glucosidic bond and transglycosylation specificity of reuteransucrase from Lactobacillus reuteri 121. FEBS J. 2006, 273, 3735–3742.
- van Leeuwen, S.S.; Kralj, S.; van Geel-Schutten, I.H.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural analysis of the α-D-glucan (EPS180) produced by the Lactobacillus reuteri strain 180 glucansucrase GTF180 enzyme. Carbohydr. Res. 2008, 343, 1237–1250.
- Rodrigues, S.; Lona, L.; Franco, T. Effect of phosphate concentration on the production of dextransucrase by Leuconostoc mesenteroides NRRL B512F. Bioprocess Biosyst. Eng. 2003, 26, 57–62.
- Otts, D.; Day, D.F. Dextransucrase secretion in Leuconostoc mesenteroides depends on the presence of a transmembrane proton gradient. J. Bacteriol. 1988, 170, 5006–5011.
- Quirasco, M.; Lopez-Munguia, A.; Remaud-Simeon, M.; Monsan, P.; Farres, A. Induction and transcription studies of the dextransucrase gene in Leuconostoc mesenteroides NRRL B-512F. Appl. Environ. Microbiol. 1999, 65, 5504–5509.
- Goyal, A.; Katiyar, S.S. Fractionation of Leuconostoc mesenteroides NRRL B-512F dextran sucrase by polyethylene glycol: A simple and effective method purification. J. Microbiol. Methods 1994, 20, 225–231.
- Purama, R.K.; Goyal, A. Identification, effective purification and functional characterization of dextransucrase from Leuconostoc mesenteroides NRRL B-640. Bioresour. Technol. 2008, 99, 3635–3642.
- Kobayashi, M.; Matsuda, K. Electrophoretic analysis of the multiple forms of dextransucrase from Leuconostoc mesenteroides. J. Biochem. 1986, 100, 615–621.
- Miller, A.W.; Eklund, S.H.; Robyt, J.F. Milligram to gram scale purification and characterization of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 1986, 147, 119–133.
- Miller, A.W.; Robyt, J.F. Stabilization of dextransucrase from Leuconostoc mesenteroides NRRL B-512F by nonionic detergents, poly(ethylene glycol) and high-molecular-weight dextran. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1984, 785, 89–96.
- Kitaoka, M.; Robyt, J.F. Use of a microtiter plate screening method for obtaining Leuconostoc mesenteroides mutants constitutive for glucansucrase. Enzym. Microb. Technol. 1998, 22, 527–531.
- Monsan, P.; Lopez, A. On the production of dextran by free and immobilized dextransucrase. Biotechnol. Bioeng. 1981, 23, 2027–2037.
- Neubauer, H.; Bauche, A.; Mollet, B. Molecular characterization and expression analysis of the dextransucrase DsrD of Leuconostoc mesenteroides Lcc4 in homologous and heterologous Lactococcus lactis cultures. Microbiology 2003, 149, 973–982.
- Robyt, J.F.; Walseth, T.F. Production, purification, and properties of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 1979, 68, 95–111.
- Miao, M.; Ma, Y.; Jiang, B.; Cui, S.W.; Jin, Z.; Zhang, T. Characterisations of Lactobacillus reuteri SK24. 003 glucansucrase: Implications for α-gluco-poly and oligosaccharides biosynthesis. Food Chem. 2017, 222, 105–112.
- Nigam, M.; Goyal, A.; Katiyar, S.S. High yield purification of dextransucrase from Leuconostoc mesenteroides NRRL B-512F by phase partitioning. J. Food Biochem. 2006, 30, 12–20.
- Amari, M.; Arango, L.; Gabriel, V.; Robert, H.; Morel, S.; Moulis, C.; Gabriel, B.; Remaud-Siméon, M.; Fontagné-Faucher, C. Characterization of a novel dextransucrase from Weissella confusa isolated from sourdough. Appl. Microbiol. Biotechnol. 2013, 97, 5413–5422.
- Wang, C.; Chen, S.; Zhang, H.B.; Li, Y.; Hu, X.Q. Characterization of the inserted mutagenesis dextransucrases from Leuconostoc mesenteroides 0326 to produce hyperbranched dextran. Int. J. Biol. Macromol. 2018, 112, 584–590.
- Kim, Y.M.; Yeon, M.J.; Choi, N.S.; Chang, Y.H.; Jung, M.Y.; Song, J.J.; Kim, J.S. Purification and characterization of a novel glucansucrase from Leuconostoc lactis EG001. Microbiol. Res. 2010, 165, 384–391.
- Rühmkorf, C.; Bork, C.; Mischnick, P.; Rübsam, H.; Becker, T.; Vogel, R.F. Identification of Lactobacillus curvatus TMW 1.624 dextransucrase and comparative characterization with Lactobacillus reuteri TMW 1.106 and Lactobacillus animalis TMW 1.971 dextransucrases. Food Microbiol. 2013, 34, 52–61.
- Vidal, R.F.; Martínez, A.; Moulis, C.; Escalier, P.; Morel, S.; Remaud-Simeon, M.; Monsan, P. A novel dextransucrase is produced by Leuconostoc citreum strain B/110-1-2: An isolate used for the industrial production of dextran and dextran derivatives. J. Ind. Microbiol. Biotechnol. 2011, 38, 1499–1506.
- Yi, A.R.; Lee, S.R.; Jang, M.U.; Park, J.M.; Eom, H.J.; Han, N.S.; Kim, T.J. Cloning of dextransucrase gene from Leuconostoc citreum HJ-P4 and its high-level expression in E. coli by low temperature induction. J. Microbiol. Biotechnol. 2009, 19, 829–835.
- Ko, J.A.; Jeong, H.J.; Ryu, Y.B.; Park, S.J.; Wee, Y.J.; Kim, D.; Kim, Y.M.; Lee, W.S. Large increase in Leuconostoc citreum KM20 dextransucrase activity achieved by changing the strain/inducer combination in an E. coli expression system. J. Microbiol. Biotechnol. 2012, 22, 510–515.
- Passerini, D.; Vuillemin, M.; Ufarté, L.; Morel, S.; Loux, V.; Fontagné-Faucher, C.; Monsan, P.; Remaud-Siméon, M.; Moulis, C. Inventory of the GH70 enzymes encoded by Leuconostoc citreum NRRL B-1299-identification of three novel α-transglucosylases. FEBS J. 2015, 282, 2115–2130.
- Monchois, V.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.M. Cloning and sequencing of a gene coding for an extracellular dextransucrase (DSRB) from Leuconostoc mesenteroides NRRL B-1299 synthesizing only a alpha (1-6) glucan. FEMS Microbiol. Lett. 1998, 159, 307–315.
- Bozonnet, S.; Dols-Laffargue, M.; Fabre, E.; Pizzut, S.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.M. Molecular characterization of DSR-E, an alpha-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 2002, 184, 5753–5761.
- Zhang, H.; Hu, Y.; Zhu, C.; Zhu, B.; Wang, Y. Cloning, sequencing and expression of a dextransucrase gene (dexYG) from Leuconostoc mesenteroides. Biotechnol. Lett. 2008, 30, 1441–1446.
- Kang, H.K.; Kim, Y.M.; Kim, D.M. Functional, genetic, and bioinformatic characterization of dextransucrase (DSRBCB4) gene in Leuconostoc mesenteroides B-1299 CB4. J. Microbiol. Biotechnol. 2008, 18, 1050–1058.
- Yoon, S.H.; Fulton, D.B.; Robyt, J.F. Enzymatic synthesis of L-DOPA alpha-glycosides by reaction with sucrose catalyzed by four different glucansucrases from four strains of Leuconostoc mesenteroides. Carbohydr. Res. 2010, 345, 1730–1735.
- Yoon, S.H.; Bruce Fulton, D.; Robyt, J.F. Enzymatic synthesis of two salicin analogues by reaction of salicyl alcohol with Bacillus macerans cyclomaltodextrin glucanyltransferase and Leuconostoc mesenteroides B-742CB dextransucrase. Carbohydr. Res. 2004, 339, 1517–1529.
- Chellapandian, M.; Larios, C.; Sanchez-Gonzalez, M.; Lopez-Munguia, A. Production and properties of a dextransucrase from Leuconostoc mesenteroides IBT-PQ isolated from ‘pulque’, a traditional Aztec alcoholic beverage. J. Ind. Microbiol. Biotechnol. 1998, 21, 51–56.
- Siddiqui, N.N.; Aman, A.; Qader, S.A. Mutational analysis and characterization of dextran synthesizing enzyme from wild and mutant strain of Leuconostoc mesenteroides. Carbohydr. Polym. 2013, 91, 209–216.
- Yalin, Y.; Jin, L.; Jianhua, W.; Da, T.; Zigang, T. Expression and characterization of dextransucrase gene dsrX from Leuconostoc mesenteroides in Escherichia coli. J. Biotechnol. 2008, 133, 505–512.
- Monchois, V.; Remaud-Simeon, M.; Russell, R.R.; Monsan, P.; Willemot, R.M. Characterization of Leuconostoc mesenteroides NRRL B-512F dextransucrase (DSRS) and identification of amino-acid residues playing a key role in enzyme activity. Appl. Microbiol. Biotechnol. 1997, 48, 465–472.
- Funane, K.; Ishii, T.; Matsushita, M.; Hori, K.; Mizuno, K.; Takahara, H.; Kitamura, Y.; Kobayashi, M. Water-soluble and water-insoluble glucans produced by Escherichia coli recombinant dextransucrases from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 2001, 334, 19–25.
- Gilmore, K.S.; Russell, R.R.; Ferretti, J.J. Analysis of the Streptococcus downei gtfS gene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect. Immun. 1990, 58, 2452–2458.
- Hanada, N.; Fukushima, K.; Nomura, Y.; Senpuku, H.; Hayakawa, M.; Mukasa, H.; Shiroza, T.; Abiko, Y. Cloning and nucleotide sequence analysis of the Streptococcus sobrinus gtfU gene that produces a highly branched water-soluble glucan. Biochim. Biophys. Acta 2002, 1570, 75–79.
- Tsumori, H.; Minami, T.; Kuramitsu, H.K. Identification of essential amino acids in the Streptococcus mutans glucosyltransferases. J. Bacteriol. 1997, 179, 3391–3396.
- Shimamura, A.; Nakano, Y.J.; Mukasa, H.; Kuramitsu, H.K. Identification of amino acid residues in Streptococcus mutans glucosyltransferases influencing the structure of the glucan product. J. Bacteriol. 1994, 176, 4845–4850.
- Bounaix, M.S.; Robert, H.; Gabriel, V.; Morel, S.; Remaud-Siméon, M.; Gabriel, B.; Fontagné-Faucher, C. Characterization of dextran-producing Weissella strains isolated from sourdoughs and evidence of constitutive dextransucrase expression. FEMS Microbiol. Lett. 2010, 311, 18–26.
- Shukla, S.; Shi, Q.; Maina, N.H.; Juvonen, M.; Goyal, A. Weissella confusa Cab3 dextransucrase: Properties and in vitro synthesis of dextran and glucooligosaccharides. Carbohydr. Polym. 2014, 101, 554–564.
- Kajala, I.; Shi, Q.; Nyyssölä, A.; Maina, N.H.; Hou, Y.; Katina, K.; Tenkanen, M.; Juvonen, R. Cloning and characterization of a Weissella confusa dextransucrase and its application in high fibre baking. PLoS ONE 2015, 10, e0116418.
- Wangpaiboon, K.; Padungros, P.; Nakapong, S.; Charoenwongpaiboon, T.; Rejzek, M.; Field, R.A.; Pichyangkura, R. An α-1,6-and α-1,3-linked glucan produced by Leuconostoc citreum ABK-1 alternansucrase with nanoparticle and film-forming properties. Sci. Rep. 2018, 8, 8340.
- Miao, M.; Ma, Y.; Jiang, B.; Huang, C.; Li, X.; Cui, S.W.; Zhang, T. Structural investigation of a neutral extracellular glucan from Lactobacillus reuteri SK24.003. Carbohydr. Polym. 2014, 106, 384–392.
- Argüello-Morales, M.A.; Remaud-Simeon, M.; Pizzut, S.; Sarçabal, P.; Willemot, R.; Monsan, P. Sequence analysis of the gene encoding alternansucrase, a sucrose glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. FEMS Microbiol. Lett. 2000, 182, 81–85.
- Kralj, S.; Stripling, E.; Sanders, P.; van Geel-Schutten, G.H.; Dijkhuizen, L. Highly hydrolytic reuteransucrase from probiotic Lactobacillus reuteri strain ATCC 55730. Appl. Environ. Microbiol. 2005, 71, 3942–3950.
- Miao, M.; Ma, Y.; Huang, C.; Jiang, B.; Cui, S.W.; Zhang, T. Physicochemical properties of a water soluble extracellular homopolysaccharide from Lactobacillus reuteri SK24.003. Carbohydr. Polym. 2015, 131, 377–383.
- Côté, G.L.; Skory, C.D. Cloning, expression, and characterization of an insoluble glucan-producing glucansucrase from Leuconostoc mesenteroides NRRL B-1118. Appl. Microbiol. Biotechnol. 2012, 93, 2387–2394.
- Shiroza, T.; Ueda, S.; Kuramitsu, H.K. Sequence analysis of the gtfB gene from Streptococcus mutans. J. Bacteriol. 1987, 169, 4263–4270.
- Ueda, S.; Shiroza, T.; Kuramitsu, H.K. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene 1988, 69, 101–109.
- Gangoiti, J.; van Leeuwen, S.S.; Gerwig, G.J.; Duboux, S.; Vafiadi, C.; Pijning, T.; Dijkhuizen, L. 4,3-α-Glucanotransferase, a novel reaction specificity in glycoside hydrolase family 70 and clan GH-H. Sci. Rep. 2017, 7, 39761.
- Dobruchowska, J.M.; Gerwig, G.J.; Kralj, S.; Grijpstra, P.; Leemhuis, H.; Dijkhuizen, L.; Kamerling, J.P. Structural characterization of linear isomalto-/malto-oligomer products synthesized by the novel GTFB 4,6-α-glucanotransferase enzyme from Lactobacillus reuteri 121. Glycobiology 2012, 22, 517–528.
- Leemhuis, H.; Dijkman, W.P.; Dobruchowska, J.M.; Pijning, T.; Grijpstra, P.; Kralj, S.; Kamerling, J.P.; Dijkhuizen, L. 4,6-α-Glucanotransferase activity occurs more widespread in Lactobacillus strains and constitutes a separate GH70 subfamily. Appl. Microbiol. Biotechnol. 2013, 97, 181–193.
- Pijning, T.; Gangoiti, J.; Te Poele, E.M.; Börner, T.; Dijkhuizen, L. Insights into Broad-Specificity Starch Modification from the Crystal Structure of Limosilactobacillus Reuteri NCC 2613 4,6-α-Glucanotransferase GtfB. J. Agric. Food Chem. 2021, 69, 13235–13245.
- Kang, H.K.; Oh, J.S.; Kim, D. Molecular characterization and expression analysis of the glucansucrase DSRWC from Weissella cibaria synthesizing a alpha (1→6) glucan. FEMS Microbiol. Lett. 2009, 292, 33–41.
- Gangoiti, J.; van Leeuwen, S.S.; Vafiadi, C.; Dijkhuizen, L. The Gram-negative bacterium Azotobacter chroococcum NCIMB 8003 employs a new glycoside hydrolase family 70 4,6-α-glucanotransferase enzyme (GtfD) to synthesize a reuteran like polymer from maltodextrins and starch. Biochim. Biophys. Acta 2016, 1860, 1224–1236.
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. The Exiguobacterium sibiricum 255-15 GtfC Enzyme Represents a Novel Glycoside Hydrolase 70 Subfamily of 4,6-α-Glucanotransferase Enzymes. Appl. Environ. Microbiol. 2016, 82, 756–766.
- Tsuchiya, H.; Koepsell, H.; Corman, J.; Bryant, G.; Bogard, M.; Feger, V.; Jackson, R. The effect of certain cultural factors on production of dextransucrase by Leuconostoc mesenteroides. J. Bacteriol. 1952, 64, 521–526.
- Du, R.; Zhao, F.; Pan, L.; Han, Y.; Xiao, H.; Zhou, Z. Optimization and purification of glucansucrase produced by Leuconostoc mesenteroides DRP2-19 isolated from Chinese Sauerkraut. Prep. Biochem. Biotechnol. 2018, 48, 465–473.
- Molina, M.; Moulis, C.; Monties, N.; Pizzut-Serin, S.; Guieysse, D.; Morel, S.; Cioci, G.; Remaud-Simeéon, M. Deciphering an undecided enzyme: Investigations of the structural determinants involved in the linkage specificity of alternansucrase. ACS Catal. 2019, 9, 2222–2237.
- Qader, S.A.U.; Aman, A.; Bano, S.; Syed, N.; Azhar, A. The effect of calcium ions and temperature on the production, activity and stability of dextransucrase from the newly isolated strain Leuconostoc mesenteroides PCSIR-4. Rom. J. Biochem. 2008, 45, 159–168.
- Bai, Y.; van der Kaaij, R.M.; Leemhuis, H.; Pijning, T.; van Leeuwen, S.S.; Jin, Z.; Dijkhuizen, L. Biochemical characterization of the Lactobacillus reuteri glycoside hydrolase family 70 GTFB type of 4, 6-α-glucanotransferase enzymes that synthesize soluble dietary starch fibers. Appl. Environ. Microbiol. 2015, 81, 7223–7232.
- Côté, G.L.; Robyt, J.F. Isolation and partial characterization of an extracellular glucansucrase from Leuconostoc mesenteroides NRRL B-1355 that synthesizes an alternating (1→6), (1→3)-α-D-glucan. Carbohydr. Res. 1982, 101, 57–74.
- Du, R.; Qiao, X.; Wang, Y.; Zhao, B.; Han, Y.; Zhou, Z. Determination of glucansucrase encoding gene in Leuconostoc mesenteroides. Int. J. Biol. Macromol. 2019, 137, 761–766.




