Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Priscilla Cascetta | -- | 2036 | 2022-11-24 04:03:08 | | | |
| 2 | Dean Liu | -3 word(s) | 2033 | 2022-11-25 03:33:39 | | | | |
| 3 | Dean Liu | + 8 word(s) | 2041 | 2022-11-28 09:08:45 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Cascetta, P.; Marinello, A.; Lazzari, C.; Gregorc, V.; Bianco, R.; Normanno, N.; Morabito, A.; Planchard, D. KRAS in Non-Small Cell Lung Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/36171 (accessed on 05 March 2026).
Cascetta P, Marinello A, Lazzari C, Gregorc V, Bianco R, Normanno N, et al. KRAS in Non-Small Cell Lung Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/36171. Accessed March 05, 2026.
Cascetta, Priscilla, Arianna Marinello, Chiara Lazzari, Vanesa Gregorc, Roberto Bianco, Nicola Normanno, Alessandro Morabito, David Planchard. "KRAS in Non-Small Cell Lung Cancer" Encyclopedia, https://encyclopedia.pub/entry/36171 (accessed March 05, 2026).
Cascetta, P., Marinello, A., Lazzari, C., Gregorc, V., Bianco, R., Normanno, N., Morabito, A., & Planchard, D. (2022, November 24). KRAS in Non-Small Cell Lung Cancer. In Encyclopedia. https://encyclopedia.pub/entry/36171
Cascetta, Priscilla, et al. "KRAS in Non-Small Cell Lung Cancer." Encyclopedia. Web. 24 November, 2022.
Copy Citation
In NSCLC (Non-Small Cell Lung Cancer), KRAS (Kirsten Rat sarcoma virus) mutations occur in up to 30% of all cases, most frequently at codon 12 and 13. KRAS mutations have been linked to adenocarcinoma histology, positive smoking history, and Caucasian ethnicity, although differences have been described across KRAS mutational variants subtypes. KRAS mutations often concur with other molecular alterations, notably TP53, STK11, and KEAP1, which could play an important role in treatment efficacy and patient outcomes.
KRAS
NSCLC
lung cancer
adagrasib
1. Introduction
With almost 1.8 million deaths per year, lung cancer remains the leading cause of cancer-related death worldwide. Non-small cell lung cancer (NSCLC) accounts for 85% of all diagnosed cases, with adenocarcinoma being the most common histological subtype [1][2]. Progress in molecular characterization and development of new targeted therapies have changed the treatment scenario of NSCLC in recent years. So far, molecular alterations thought to be undruggable for many years, such as KRAS mutations, may now benefit from targeted agents.
KRAS (Kirsten Rat sarcoma virus) proteins are small GTP-ases belonging to the wider family of RAS proteins. The KRAS gene is located in the short arm of chromosome 12 (12 p) and its transcripts may further undergo an alternative splicing process, resulting in two distinct proteins of approximately 21 KDa each: KRAS 4A and KRAS 4B [3]. Even though differences in physiological expression have been reported in some preclinical data [4], little is known about the real impact of these alternatively spliced variants in NSCLC.
Structurally, all RAS isoforms have a highly conserved catalytic region, implied in GTP bounding and hydrolysis. Differences across RAS isoforms are mainly founded at the C-terminal hyper variable region (HVR) domain, responsible for post-transcriptional modifications and interaction with the phospholipidic membrane bilayer [5][6].
In physiological conditions, KRAS proteins are key elements in transducing signaling from activated receptor tyrosine kinase (RTK) to downstream intracellular pathways. Indeed, intracellular KRAS proteins may present in two distinct functional forms: KRAS-GTP and KRAS-GDP, which correspond to “active” and “inactive” states, respectively. Transition from the inactive state (KRAS-GDP) to active state (KRAS-GTP) is usually mediated by the guanine nucleotide exchange factors (GEFs) via the releasing of guanosine diphosphate (GDP) and the subsequent binding of guanosine triphosphate (GTP). Many GEFs proteins have been identified to date, with the GEF Son of Sevenless (SOS1) playing a crucial role in KRAS activation. Of note, this process determines key structural modification in KRAS proteins mainly involving the switch pockets I and II, which serve as the binding interface for effector proteins [7][8]. However, the transition from the inactive to active form is complex and usually requires other proteins (e.g., the adaptor protein Grb2 and the SOS1/Grb2 recruiter SHP-2) which directly interact with upstream phosphorylated RTKs [9]. Contrarily, transition from the KRAS-GTP to KRAS-GDP binding state is facilitated by GTPase activating proteins (GAPs) such as NF1, which accelerate the hydrolysis of GTP to GDP [10].
Downstream pathways of KRAS normally include the RAF-MEK-ERK mitogen-activated protein kinase (MAPK) pathway, the phosphatidylinositol 3-kinase (PI3K)-AKT-mTOR, and the RAS-like (RAL) pathways [5], whose deregulations have been linked with abnormal cellular proliferation, migration, and death (Figure 1).
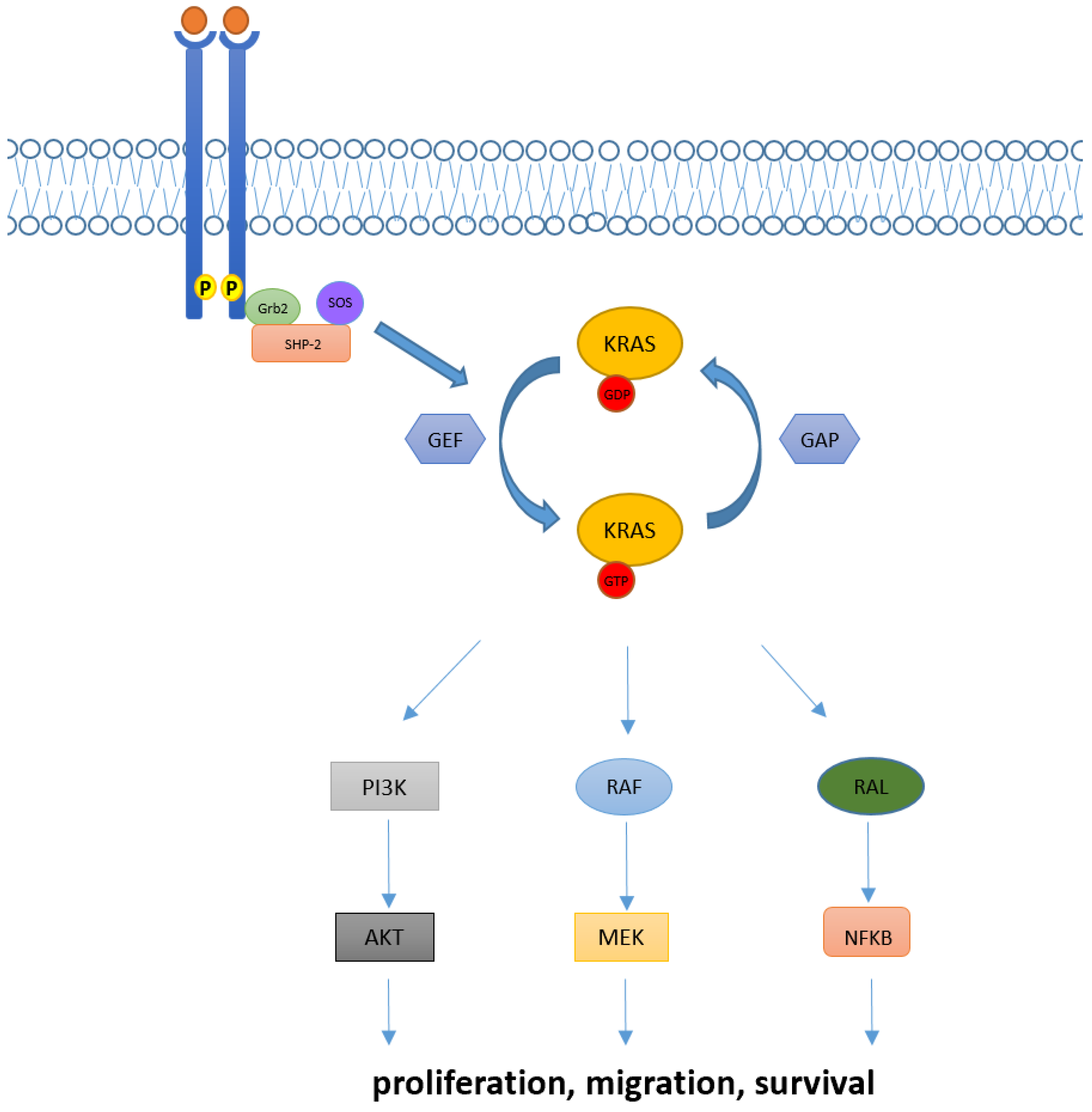
Figure 1. KRAS activation and its downstream pathway. KRAS proteins are key elements in transducing extracellular signaling to downstream pathways. Intracellular KRAS proteins may present in two functional forms, KRAS-GTP and KRAS-GDP, which correspond to their active and inactive state, respectively. Transition from the KRAS-GDP to KRAS-GTP binding state is usually mediated by GEFs proteins and occurs via other adaptor and recruiter proteins, such as Grb2 and SHP-2. Conversely, transition from KRAS-GDP to KRAS-GTP usually requires GAPs proteins. Downstream signaling of KRAS normally includes RAF-MEK-ERK, PI3K/AKT and RAL pathways, implied in the regulation of cellular proliferation, migration, and death.
The researchers provides an overview of the biology and clinic-pathological characteristics of KRAS mutant NSCLC patients. Moreover, researchers discuss therapeutic strategies that have been explored in these subgroups of patients and the clinical data leading to the approval of sotorasib and adagrasib, the first compounds that efficiently inhibit KRAS G12C mutation. Researchers then focus on acquired resistance mechanisms evidenced to date and researchers further assess the role of immune checkpoint inhibitors in KRAS mutant patients. Finally, researchers look into ongoing trials and new KRAS inhibitors, which are currently being tested.
2. Techniques for Detection of KRAS Mutations
The assessment of the KRAS status in patients affected by colorectal cancer and NSCLC is mandatory for a correct therapeutical management [11]. The main techniques allowing KRAS point mutations are polymerase chain reaction (PCR) based methods, such as high-resolution melting and real time RCP (RT.-PCR), and next generation sequencing (NGS) [12][13][14]. RT-PCR allows the highly sensitive qualitative detection and identification of a fixed number of mutations in specific areas of the gene [13]. Highly verified assays, such as Cobas Mutation Test V2, reaches a 100% level of accuracy in tissue samples, analyzing exons 2, 3, and 4 of the KRAS gene [15]. NGS technologies, being DNA-based or RNA-based, enable the simultaneous analysis of a high variety of genes and loci, at the price of reducing sensitivity, when compared to PCR [16]. With the advent of new molecular targets, an exhaustive characterization of lung cancer patients has become essential, making NGS an extremely valuable tool [17][18][19][20].
The sensitivity of KRAS mutation detection has been evaluated and compared in various types of specimens, such as tissue, plasma, and other body fluids, finding a high rate of concordance between tissue and liquid biopsy both for PCR and NGS [21][22]. The use of liquid biopsy is of great relevance for the management of advanced NSCLC, enabling the identification of KRAS mutations at the baseline, but also to picture disease heterogeneity, to evaluate response to treatment in time and to identify molecular mechanisms of resistance to targeted molecules [22][23][24]. Overall, tissue-based testing remains the preferred test for most cancer patients, although liquid biopsy may be chosen when faster results will be clinically important, or when obtaining tissue specimens is not possible or inappropriate [23] However, it is important to point out that reimbursement for broad coverage is still limited [25].
3. KRAS in NSCLC: Different Alterations and Patients’ Characteristics
In NSCLC, KRAS mutations are founded in up to 30% of all cases and most frequently involve the codon 12 (90% of cases), while less common mutations are observed in codon 13 (2–6%) and 61 (1%) [26][27]. Within codon 12 mutations, the one resulting in the substitution of glycine with cysteine (KRAS G12C) represents the most prevalent KRAS alteration in NSCLC (40% of all KRAS mutant cases), followed by substitution of glycine with valine (KRAS G12V, 21%), and substitution of glycine with aspartic acid (KRAS G12D, 17%). Indeed, other point mutation in codon 12 such as G12A/R/S are rare [26][27][28]. Functionally, mutations at codon 12 and 13 result in reduced GAP-mediated hydrolysis via allosteric bloc that hinders the GTP-ase site of KRAS, whereas alterations in codon 61 abolish both intrinsic and GAP-mediated GTP hydrolysis of KRAS [29]. Of note, KRAS mutant variants can differently activate their downstream pathway. Indeed, the hydrophobic G12C and G12V preferentially activate the RAL pathway, while the hydrophilic G12D mainly acts via PI3K-AKT signaling. KRAS G12C weakens the interaction with PI3K, and the bulky aspartic acid in the G12D prevents the formation of the binding with RelA/RelB [30]. Besides NSCLC, KRAS mutations are also frequent in other tumor types, such as pancreatic and colorectal adenocarcinoma (88% and 50% of all cases, respectively) [28][31], with KRAS G12D mutations being the most common alteration among these histotypes (32% and 46% of all cases, respectively) [32].
In lung cancer patients, KRAS mutations have been associated with Caucasian ethnicity (Caucasian vs. Asian population: 26% vs. 11%), female sex (female vs. males: 31.35 vs. 23.7%, p < 0.0001), adenocarcinoma histology (adenocarcinoma vs. squamous cell carcinoma: 37.2% vs. 4.4%), and a positive smoking history (smokers vs. non-smokers: 30% vs. 11%) [26][28]. However, these characteristics may strongly depend on single mutant KRAS variants. As such, KRAS G12C/G12A/G12V/G13C mutations are more frequently found in current or former smokers, while prevalence of KRAS G12D/G12S/G13D mutations is higher among non-smokers [26][33]. Furthermore, the median age for KRAS G12C patients seems to be significantly lower than that observed for other KRAS mutants (KRAS G12C vs. other KRAS-MUT: 63.1 vs. 65.9 years, p = 0.0092) [34], although this evidence has not been further confirmed in other reports [26]. Finally, the metastatic pattern could also slightly differ across single KRAS mutant variants, since KRAS G12C mutants are more likely associated with lung metastasis (KRAS G12C vs. non–G12C: 38% vs. 21%; p = 0.043) and less associated with either pleural metastasis or lymphangitic carcinomatosis (KRAS G12C vs. non–G12C: 4% vs. 39%, p = 0.0001) [35]. Of note, KRAS mutant patients do not have a higher brain tropism than those without KRAS alterations, and the same incidence of brain metastasis has been observed across different KRAS mutant variants (KRAS mut vs. KRAS wild-type: 33% vs. 40%, p = 0.17; KRAS G12C vs. KRAS other: 40% vs. 41%, p = 0.74) [36].
4. Predictive and Prognostic Role of KRAS Mutations in NSCLC Patients
Multiple studies have evaluated the predictive and prognostic role of KRAS mutations in NSCLC.
In an early setting, different reports have demonstrated similar overall survival (OS) and disease-free survival (DFS) in KRAS mutants versus wild type patients (OS HR = 1.17, DFS HR = 1.04) [37][38]. However, these results were not further confirmed and other reports eventually demonstrated a worse OS and DFS for KRAS mutant patients (DFS p = 0.038, OS HR = 1.30; p = 0.002) [37].
It has been asked if the prognostic and predictive significance of KRAS mutation could depend on specific variants. Indeed, Finn et al. recently demonstrated a negative prognostic impact for KRAS G12C completely resected patients compared to both the KRAS wild type and other mutant variants (OS KRAS G12C versus other KRAS HR = 1.39, p = 0.031; KRAS G12C versus no KRAS HR = 1.32, p = 0.028) [34]. Contrarily, no prognostic nor predictive value for codon 12 mutations has been identified in a pooled meta-analysis conducted in early stage disease, while codon 13 mutations have been associated with a possible deleterious effect to adjuvant chemotherapy [37].
In an advanced setting, a meta-analysis of 43 selected studies demonstrated an impaired OS and progression-free survival (PFS) for KRAS mutants (OS HR = 1.71; 95% CI [1.07, 2.84]; PFS HR = 1.18; 95% CI [1.02, 1.36]). However, these results were only observed when considering observational studies and not further confirmed in randomized clinical trials (OS HR = 1.10; 95% CI [0.88–1.38]; PFS HR = 1.03; 95% CI [0.80–1.33]) [39]. Likewise, discordant results about the prognostic and predictive role of KRAS mutations have been reported in the advanced setting [27][40][41]. Furthermore, no differences in OS have been observed across different KRAS mutations, although a better response to taxanes has been reported for KRAS G12V mutants [33][42].
In conclusion, the prognostic and predictive role of KRAS mutations in NSCLC is still a matter of debate and discordant results provided so far need further assessment.
5. Concurrent Molecular Alterations in KRAS Positive NSCLC Patients
Concurrent molecular alterations are often found in KRAS mutant NSCLC patients. In the largest cohort available to date, Riely et al. described a median of seven concomitant mutations in KRAS positive NSCLC patients, the most frequent being TP53 (39%), STK11 (30%), KEAP1 (24%), RBM10 (15%), and PTPRD (15%). Curiously, those harboring concomitant mutations in either STK11 or KEAP1 had a shorter OS (STK11: HR = 2, p = 0.006; KEAP1: HR = 2.8, p < 0.001) and the negative prognostic impact of STK11 and KEAP1 co-alterations has been further confirmed in multiple studies [43][44]. Of note, neither the TP53 status nor number of concurrent mutations affected OS [45]. In another retrospective study, Passaro et al. demonstrated that 77% of patients with KRAS mutations had a concurrent molecular alteration. Interestingly, the percentage of concomitant alterations did not differ between G12C and non G-12C KRAS mutants. Again, the most frequently reported co-alteration was TP53 (23%) followed by STK11 (15%). Importantly, alterations in CDK2/4 and receptor tyrosine-kinase (RTK) have also been described in KRAS mutant cases [46].
Importantly, concurrent alterations might affect the tumor microenvironment with consequent alterations in the immunogenic phenotype. As such, Skoulidis et al. demonstrated a lower immunogenic infiltrate in STK11 positive tumors, while the opposite was seen for concomitant TP53-mutated tumors [47].
It has also been demonstrated that co-mutations could differentially activate downstream pathways. Indeed, engineered mice harboring KRAS G12D alone or with concomitant TP53 or STK11 mutations, respectively, hyper-activated MEK/ERK or AKT/SRC pathways [48].
Concurrent alterations with other key oncogenic drivers have also been described. Of note, KRAS mutations are found in approximately 3% of MET ex14 alterations [49][50], and a few cases of concomitant KRAS/ALK and KRAS/ROS1 alterations have been reported [51][52]. Interestingly, KRAS mutations have been found as mechanisms of resistance to the first, second, and third generation of EGFR TKI [53][54][55][56].
References
- Ganti, A.K.; Klein, A.B.; Cotarla, I.; Seal, B.; Chou, E. Update of Incidence, Prevalence, Survival, and Initial Treatment in Patients With Non–Small Cell Lung Cancer in the US. JAMA Oncol. 2021, 7, 1824–1832.
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung Cancer. Lancet 2021, 398, 535–554.
- KRAS—An Overview|ScienceDirect Topics. Available online: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/kras (accessed on 31 July 2022).
- Plowman, S.J.; Berry, R.L.; Bader, S.A.; Luo, F.; Arends, M.J.; Harrison, D.J.; Hooper, M.L.; Patek, C.E. K-Ras 4A and 4B Are Co-Expressed Widely in Human Tissues, and Their Ratio Is Altered in Sporadic Colorectal Cancer. J. Exp. Clin. Cancer Res. 2006, 25, 259–267.
- Hancock, J.F. Ras Proteins: Different Signals from Different Locations. Nat. Rev. Mol. Cell Biol. 2003, 4, 373–385.
- Westcott, P.M.K.; To, M.D. The Genetics and Biology of KRAS in Lung Cancer. Chin. J. Cancer 2013, 32, 63–70.
- Pantsar, T. The Current Understanding of KRAS Protein Structure and Dynamics. Comput. Struct. Biotechnol. J. 2019, 18, 189–198.
- Drugging an Undruggable Pocket on KRAS|PNAS. Available online: https://www.pnas.org/doi/10.1073/pnas.1904529116 (accessed on 22 September 2022).
- Dance, M.; Montagner, A.; Salles, J.-P.; Yart, A.; Raynal, P. The Molecular Functions of Shp2 in the Ras/Mitogen-Activated Protein Kinase (ERK1/2) Pathway. Cell. Signal. 2008, 20, 453–459.
- Maertens, O.; Cichowski, K. An Expanding Role for RAS GTPase Activating Proteins (RAS GAPs) in Cancer. Adv. Biol. Regul. 2014, 55, 1–14.
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the Use of Next-Generation Sequencing (NGS) for Patients with Metastatic Cancers: A Report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505.
- Gundry, C.N.; Vandersteen, J.G.; Reed, G.H.; Pryor, R.J.; Chen, J.; Wittwer, C.T. Amplicon Melting Analysis with Labeled Primers: A Closed-Tube Method for Differentiating Homozygotes and Heterozygotes. Clin. Chem. 2003, 49, 396–406.
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real Time Quantitative PCR. Genome Res. 1996, 6, 986–994.
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238.
- Li, J.; Gan, S.; Blair, A.; Min, K.; Rehage, T.; Hoeppner, C.; Halait, H.; Brophy, V.H. A Highly Verified Assay for KRAS Mutation Detection in Tissue and Plasma of Lung, Colorectal, and Pancreatic Cancer. Arch. Pathol. Lab. Med. 2019, 143, 183–189.
- Elazezy, M.; Joosse, S.A. Techniques of Using Circulating Tumor DNA as a Liquid Biopsy Component in Cancer Management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378.
- Gagan, J.; Van Allen, E.M. Next-Generation Sequencing to Guide Cancer Therapy. Genome Med. 2015, 7, 80.
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; Van Schil, P.E.; Hellmann, M.D.; et al. Metastatic Non-Small Cell Lung Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2018, 29, iv192–iv237.
- Hagemann, I.S.; Devarakonda, S.; Lockwood, C.M.; Spencer, D.H.; Guebert, K.; Bredemeyer, A.J.; Al-Kateb, H.; Nguyen, T.T.; Duncavage, E.J.; Cottrell, C.E.; et al. Clinical Next-Generation Sequencing in Patients with Non-Small Cell Lung Cancer. Cancer 2015, 121, 631–639.
- Tsoulos, N.; Papadopoulou, E.; Metaxa-Mariatou, V.; Tsaousis, G.; Efstathiadou, C.; Tounta, G.; Scapeti, A.; Bourkoula, E.; Zarogoulidis, P.; Pentheroudakis, G.; et al. Tumor Molecular Profiling of NSCLC Patients Using next Generation Sequencing. Oncol. Rep. 2017, 38, 3419–3429.
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180.
- Horn, L.; Whisenant, J.G.; Wakelee, H.; Reckamp, K.L.; Qiao, H.; Leal, T.A.; Du, L.; Hernandez, J.; Huang, V.; Blumenschein, G.R.; et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients With ALK+ Lung Cancer. J. Thorac. Oncol. 2019, 14, 1901–1911.
- Pascual, J.; Attard, G.; Bidard, F.-C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO Recommendations on the Use of Circulating Tumour DNA Assays for Patients with Cancer: A Report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768.
- Heitzer, E.; van den Broek, D.; Denis, M.G.; Hofman, P.; Hubank, M.; Mouliere, F.; Paz-Ares, L.; Schuuring, E.; Sültmann, H.; Vainer, G.; et al. Recommendations for a Practical Implementation of Circulating Tumor DNA Mutation Testing in Metastatic Non-Small-Cell Lung Cancer. ESMO Open 2022, 7, 100399.
- IJzerman, M.J.; de Boer, J.; Azad, A.; Degeling, K.; Geoghegan, J.; Hewitt, C.; Hollande, F.; Lee, B.; To, Y.H.; Tothill, R.W.; et al. Towards Routine Implementation of Liquid Biopsies in Cancer Management: It Is Always Too Early, until Suddenly It Is Too Late. Diagnostics 2021, 11, 103.
- Judd, J.; Abdel Karim, N.; Khan, H.; Naqash, A.R.; Baca, Y.; Xiu, J.; VanderWalde, A.M.; Mamdani, H.; Raez, L.E.; Nagasaka, M.; et al. Characterization of KRAS Mutation Subtypes in Non–Small Cell Lung Cancer. Mol. Cancer Ther. 2021, 20, 2577–2584.
- Wood, K.; Hensing, T.; Malik, R.; Salgia, R. Prognostic and Predictive Value in KRAS in Non–Small-Cell Lung Cancer: A Review. JAMA Oncol. 2016, 2, 805–812.
- Reck, M.; Carbone, D.P.; Garassino, M.; Barlesi, F. Targeting KRAS in Non-Small-Cell Lung Cancer: Recent Progress and New Approaches. Ann. Oncol. 2021, 32, 1101–1110.
- Kwan, A.K.; Piazza, G.A.; Keeton, A.B.; Leite, C.A. The Path to the Clinic: A Comprehensive Review on Direct KRASG12C Inhibitors. J. Exp. Clin. Cancer Res. 2022, 41, 27.
- Ihle, N.T.; Byers, L.A.; Kim, E.S.; Saintigny, P.; Lee, J.J.; Blumenschein, G.R.; Tsao, A.; Liu, S.; Larsen, J.E.; Wang, J.; et al. Effect of KRAS Oncogene Substitutions on Protein Behavior: Implications for Signaling and Clinical Outcome. J. Natl. Cancer Inst. 2012, 104, 228–239.
- Désage, A.-L.; Léonce, C.; Swalduz, A.; Ortiz-Cuaran, S. Targeting KRAS Mutant in Non-Small Cell Lung Cancer: Novel Insights Into Therapeutic Strategies. Front. Oncol. 2022, 12, 796832.
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS Isoforms and Mutations in Cancer at a Glance. J. Cell Sci. 2016, 129, 1287–1292.
- Ruppert, A.-M.; Beau-Faller, M.; Debieuvre, D.; Ouafik, L.; Westeel, V.; Rouquette, I.; Mazières, J.; Bringuier, P.-P.; Monnet, I.; Escande, F.; et al. Outcomes of Patients With Advanced NSCLC From the Intergroupe Francophone de Cancérologie Thoracique Biomarkers France Study by KRAS Mutation Subtypes. JTO Clin. Res. Rep. 2020, 1, 100052.
- Finn, S.P.; Addeo, A.; Dafni, U.; Thunnissen, E.; Bubendorf, L.; Madsen, L.B.; Biernat, W.; Verbeken, E.; Hernandez-Losa, J.; Marchetti, A.; et al. Prognostic Impact of KRAS G12C Mutation in Patients With NSCLC: Results From the European Thoracic Oncology Platform Lungscape Project. J. Thorac. Oncol. 2021, 16, 990–1002.
- Wu, M.Y.; Zhang, E.W.; Strickland, M.R.; Mendoza, D.P.; Lipkin, L.; Lennerz, J.K.; Gainor, J.F.; Heist, R.S.; Digumarthy, S.R. Clinical and Imaging Features of Non-Small Cell Lung Cancer with G12C KRAS Mutation. Cancers 2021, 13, 3572.
- Cui, W.; Franchini, F.; Alexander, M.; Officer, A.; Wong, H.-L.; IJzerman, M.; Desai, J.; Solomon, B.J. Real World Outcomes in KRAS G12C Mutation Positive Non-Small Cell Lung Cancer. Lung Cancer 2020, 146, 310–317.
- Yu, H.A.; Sima, C.S.; Shen, R.; Kass, S.; Gainor, J.; Shaw, A.; Hames, M.; Iams, W.; Aston, J.; Lovly, C.M.; et al. Prognostic Impact of KRAS Mutation Subtypes in 677 Patients with Metastatic Lung Adenocarcinomas. J. Thorac. Oncol. 2015, 10, 431–437.
- Shepherd, F.A.; Lacas, B.; Le Teuff, G.; Hainaut, P.; Jänne, P.A.; Pignon, J.-P.; Le Chevalier, T.; Seymour, L.; Douillard, J.-Y.; Graziano, S.; et al. Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J. Clin. Oncol. 2017, 35, 2018–2027.
- Goulding, R.E.; Chenoweth, M.; Carter, G.C.; Boye, M.E.; Sheffield, K.M.; John, W.J.; Leusch, M.S.; Muehlenbein, C.E.; Li, L.; Jen, M.-H.; et al. KRAS Mutation as a Prognostic Factor and Predictive Factor in Advanced/Metastatic Non-Small Cell Lung Cancer: A Systematic Literature Review and Meta-Analysis. Cancer Treat. Res. Commun. 2020, 24, 100200.
- Rulli, E.; Marabese, M.; Torri, V.; Farina, G.; Veronese, S.; Bettini, A.; Longo, F.; Moscetti, L.; Ganzinelli, M.; Lauricella, C.; et al. Value of KRAS as Prognostic or Predictive Marker in NSCLC: Results from the TAILOR Trial. Ann. Oncol. 2015, 26, 2079–2084.
- Svaton, M.; Fiala, O.; Pesek, M.; Bortlicek, Z.; Minarik, M.; Benesova, L.; Topolcan, O. The Prognostic Role of KRAS Mutation in Patients with Advanced NSCLC Treated with Second- or Third-Line Chemotherapy. Anticancer Res. 2016, 36, 1077–1082.
- Mellema, W.W.; Masen-Poos, L.; Smit, E.F.; Hendriks, L.E.L.; Aerts, J.G.; Termeer, A.; Goosens, M.J.; Smit, H.J.M.; van den Heuvel, M.M.; van der Wekken, A.J.; et al. Comparison of Clinical Outcome after First-Line Platinum-Based Chemotherapy in Different Types of KRAS Mutated Advanced Non-Small-Cell Lung Cancer. Lung Cancer 2015, 90, 249–254.
- Arbour, K.C.; Jordan, E.; Kim, H.R.; Dienstag, J.; Yu, H.A.; Sanchez-Vega, F.; Lito, P.; Berger, M.; Solit, D.B.; Hellmann, M.; et al. Effects of Co-Occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 334–340.
- La Fleur, L.; Falk-Sörqvist, E.; Smeds, P.; Berglund, A.; Sundström, M.; Mattsson, J.S.; Brandén, E.; Koyi, H.; Isaksson, J.; Brunnström, H.; et al. Mutation Patterns in a Population-Based Non-Small Cell Lung Cancer Cohort and Prognostic Impact of Concomitant Mutations in KRAS and TP53 or STK11. Lung Cancer 2019, 130, 50–58.
- Riely, G.J.; Jordan, E.; Kim, H.R.; Yu, H.A.; Berger, M.F.; Solit, D.B.; Kris, M.G.; Ni, A.; Arcila, M.E.; Ladanyi, M. Association of Outcomes and Co-Occuring Genomic Alterations in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 9019.
- Passaro, A.; Attili, I.; Rappa, A.; Vacirca, D.; Ranghiero, A.; Fumagalli, C.; Guarize, J.; Spaggiari, L.; de Marinis, F.; Barberis, M.; et al. Genomic Characterization of Concurrent Alterations in Non-Small Cell Lung Cancer (NSCLC) Harboring Actionable Mutations. Cancers 2021, 13, 2172.
- Skoulidis, F.; Byers, L.A.; Diao, L.; Papadimitrakopoulou, V.A.; Tong, P.; Izzo, J.; Behrens, C.; Kadara, H.; Parra, E.R.; Canales, J.R.; et al. Co-Occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discov. 2015, 5, 860–877.
- Mahoney, C.L.; Choudhury, B.; Davies, H.; Edkins, S.; Greenman, C.; van Haaften, G.; Mironenko, T.; Santarius, T.; Stevens, C.; Stratton, M.R.; et al. LKB1/KRAS Mutant Lung Cancers Constitute a Genetic Subset of NSCLC with Increased Sensitivity to MAPK and MTOR Signalling Inhibition. Br. J. Cancer 2009, 100, 370–375.
- Schrock, A.B.; Frampton, G.M.; Suh, J.; Chalmers, Z.R.; Rosenzweig, M.; Erlich, R.L.; Halmos, B.; Goldman, J.; Forde, P.; Leuenberger, K.; et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J. Thorac. Oncol. 2016, 11, 1493–1502.
- Suzawa, K.; Offin, M.; Lu, D.; Kurzatkowski, C.; Vojnic, M.; Smith, R.S.; Sabari, J.K.; Tai, H.; Mattar, M.; Khodos, I.; et al. Activation of KRAS Mediates Resistance to Targeted Therapy in MET Exon 14-Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 1248–1260.
- Zhu, Y.; Lin, X.; Li, X.; Wu, L.; Chen, H.; Wang, W.; Xu, C.; Shen, J.; Wei, J.; Du, K. Concurrent ROS1 Gene Rearrangement and KRAS Mutation in Lung Adenocarcinoma: A Case Report and Literature Review. Thorac. Cancer 2018, 9, 159–163.
- Schmid, S.; Gautschi, O.; Rothschild, S.; Mark, M.; Froesch, P.; Klingbiel, D.; Reichegger, H.; Jochum, W.; Diebold, J.; Früh, M. Clinical Outcome of ALK-Positive Non–Small Cell Lung Cancer (NSCLC) Patients with De Novo EGFR or KRAS Co-Mutations Receiving Tyrosine Kinase Inhibitors (TKIs). J. Thorac. Oncol. 2017, 12, 681–688.
- Chabon, J.J.; Simmons, A.D.; Lovejoy, A.F.; Esfahani, M.S.; Newman, A.M.; Haringsma, H.J.; Kurtz, D.M.; Stehr, H.; Scherer, F.; Karlovich, C.A.; et al. Circulating Tumour DNA Profiling Reveals Heterogeneity of EGFR Inhibitor Resistance Mechanisms in Lung Cancer Patients. Nat. Commun. 2016, 7, 11815.
- Rachiglio, A.M.; Fenizia, F.; Piccirillo, M.C.; Galetta, D.; Crinò, L.; Vincenzi, B.; Barletta, E.; Pinto, C.; Ferraù, F.; Lambiase, M.; et al. The Presence of Concomitant Mutations Affects the Activity of EGFR Tyrosine Kinase Inhibitors in EGFR-Mutant Non-Small Cell Lung Cancer (NSCLC) Patients. Cancers 2019, 11, 341.
- Xiu, W.; Zhang, Q.; Yu, M.; Huang, Y.; Huang, M. Case Report: Outcome of Osimertinib Treatment in Lung Adenocarcinoma Patients With Acquired KRAS Mutations. Front. Oncol. 2021, 11, 630256.
- Yamaoka, T.; Ohmori, T.; Ohba, M.; Arata, S.; Murata, Y.; Kusumoto, S.; Ando, K.; Ishida, H.; Ohnishi, T.; Sasaki, Y. Distinct Afatinib Resistance Mechanisms Identified in Lung Adenocarcinoma Harboring an EGFR Mutation. Mol. Cancer Res. 2017, 15, 915–928.
More
Information
Subjects:
Medicine, General & Internal
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.5K
Revisions:
3 times
(View History)
Update Date:
02 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





