Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sohail Mumtaz | -- | 1799 | 2022-11-22 10:24:09 | | | |
| 2 | Conner Chen | Meta information modification | 1799 | 2022-11-23 07:16:04 | | | | |
| 3 | Conner Chen | -11 word(s) | 1788 | 2022-11-25 01:25:01 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Han, I.; Mumtaz, S.; Ashokkumar, S.; Yadav, D.K.; Choi, E.H. Virion Structure of SARS-CoV-2 and Viral Inflammation. Encyclopedia. Available online: https://encyclopedia.pub/entry/35823 (accessed on 08 February 2026).
Han I, Mumtaz S, Ashokkumar S, Yadav DK, Choi EH. Virion Structure of SARS-CoV-2 and Viral Inflammation. Encyclopedia. Available at: https://encyclopedia.pub/entry/35823. Accessed February 08, 2026.
Han, Ihn, Sohail Mumtaz, Sekar Ashokkumar, Dharmendra Kumar Yadav, Eun Ha Choi. "Virion Structure of SARS-CoV-2 and Viral Inflammation" Encyclopedia, https://encyclopedia.pub/entry/35823 (accessed February 08, 2026).
Han, I., Mumtaz, S., Ashokkumar, S., Yadav, D.K., & Choi, E.H. (2022, November 22). Virion Structure of SARS-CoV-2 and Viral Inflammation. In Encyclopedia. https://encyclopedia.pub/entry/35823
Han, Ihn, et al. "Virion Structure of SARS-CoV-2 and Viral Inflammation." Encyclopedia. Web. 22 November, 2022.
Copy Citation
COVID-19 is an epidemic infection created by SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2). SARS-CoV-2 has a single-stranded, positive-sense RNA genome with 29,891 nucleotides and 38% G + C content, encoding 9860 amino acids. Human coronaviral inflammation induces the clinical symptoms of fever, cough, and shortness of breath.
nonthermal plasma
COVID19
SARS-CoV-2
1. Introduction
COVID-19 is an epidemic infection created by SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2), and it was initially spotted in Wuhan, Hubei province, China. This infection spread worldwide and was responsible for the last pandemic. In the past 72 years (specifically 1960, 1967, 2003, 2004, 2005, 2012, and 2019), seven types of viral infections have been identified. In the 1960s, the virus infected the respiratory tract and produced a common cold, but now these infections have created major impacts all over the world. In 1965, the human coronavirus (HCoV) B814 strain was isolated from the common cold, but this strain could not be propagated in tissue culture in a laboratory.
The HCoV-229E prototypic strains (named after a student specimen coded 229E) were isolated from human respiratory tract tissue infected with the common cold [1]. In 1967, HCoV-OC43 (organ culture 43) was isolated from a tracheal organ culture [2], and both strains induced the same clinical symptoms (fever, cough, nasal discharge, sneezing, sore throat, and shortness of breath). Severe acute respiratory syndrome coronavirus (SARS-CoV) induced atypical pneumonia in humans, and it primarily appeared in Foshan City, Guangdong province, China, in November 2002. HCoV was usually harmless until the spread of SARS-CoV. HCoV-NL63 (The Netherlands 63) was obtained from the aspirate of a 7-month-old infant with bronchiolitis in The Netherlands in 2004 [3], whereas human coronavirus-Hong Kong University 1 (HCoV-HKU1) was isolated from a 71-year-old man who had been infected with pneumonia in a Hong Kong hospital in 2005 [4]. After the SARS-CoV outbreak, the Middle East respiratory syndrome (MERS) produced a pandemic in 2012. This virus was isolated in the lung sputum samples of a 60-year-old man infected with fatal pneumonia and acute renal failure [5]. The severe acute respiratory disease COVID-19 was reported in the Hubei Province of China. This infection spread to other countries and promoted severe pneumonia in a worldwide pandemic [6]. The coronavirus generally causes mild to moderate upper-respiratory tract illness; additionally, it is associated with the common cold, fever, cough, and pneumonia. From epidemiological investigation, people know these viruses are spread worldwide in a seasonal pattern [7].
The emergence of new variants of viruses induces new types of infections; with these differences, particularly genetic variations in the spike protein, the infectivity, antigenicity, and/or vaccine efficacy of the pathogen are altered. The world health organization (WHO) classifies coronavirus variants into the following: variants of concern (VOCs), variants of interest (VOIs), variants being monitored (VBMs), and variants of high consequence (VOHCs) (see www.who.int). Previously virus VOCs included Alpha, Beta, Gamma, and Delta, and now include Omicron (B.1.1.529). The 2020 coronavirus variant Alpha was detected in the UK in September, Beta in South Africa in May, Gamma in Brazil in November, and Delta in India in October. Omicron genomic sequencing indicated that 60 different genetic mutations, compared with the original SARS-CoV-2, had a fast infection rate when compared with other variants and also had fewer effects.
2. Classifications
The tenth International Committee on Taxonomy of Viruses (ICTV) assessed virus taxonomy. Coronavirus (CoV) is categorized as follows: CoV belonging to Riboviria Realm, Orthornavirae Kingdom, Pisuviricota Phylum, Pisoniviriceses Class, Nidovirales Order, Cornidovirineae Suborder, and Orthocoronavirinae are phylogenetically subdivided into four categories: (i) αCoV (HCoV-229E, HCoV-NL63), (ii) βCoV (HCoV-OC43, HCoV-HKU1, MERS-CoV, SARS-CoV, SARS CoV-2), (iii) γCoV (Avian-CoV, Duck CoV-2714), (iv) δCoV (CoV-HKU15, White CoV-HKU16). αCoV and βCoV are found in mammals; γCoV and δCoV are found in birds. The human coronaviruses envelop positive-sense, single-strand-RNA viruses and genomes 29–32 kb larger than other RNA viruses. It is a natural origin of all the human coronavirus [8].
3. Virion Structure
SARS-CoV-2 has a single-stranded, positive-sense RNA genome with 29,891 nucleotides and 38% G + C content, encoding 9860 amino acids. SARS-CoV-2 has a variable number of open reading frames (ORF); these types of fragments are expressed as a nonstructural protein (3-chymotrypsin-like protease (3CLPro), papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRp), and structural protein). The structural proteins are mainly the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. Between these, the ribonucleoprotein (N) capsid covers the E, M, and outer surface S [9][10], and protein characteristics are similar to other βHCoV.
The HCoV-SARS-CoV-2 molecular characterization of the genomic structure was reviewed by [11][12]. The SARS-CoV-2 genome encoded two flanking untranslated regions (UTRs) and a single long open reading frame encoding a polyprotein, and the genome is organized in the order of 5′-replicase (ORF1/ab)-structural proteins (spike protein (S), envelope protein (E), membrane protein (M), nucleocapsid protein (N))-3′ and also the hemagglutinin-esterase (HE) gene, which is identified as βCoVs (HCoV-OC43 and HCoV-HKU1). Two-thirds of the viral genomic region (20 kb) and the head contains open reading frame 1a and ab (ORF1a, ab), which potentially encode the nonstructural proteins (NSPs) referred to as the pp1a (NSPs1 to NSPs11) and pp1ab (NSPs12 to NSPs16) polyproteins, and the remaining 10 kb region preceding 3′-end encodes various structural proteins (S, E, M, and N). In addition, the structural genes encode nine accessory proteins: the ORF3a, ORF3d, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF14, and ORF10 genes [13].
4. Viral Inflammation
Human coronaviral inflammation induces the clinical symptoms of fever, cough, and shortness of breath. These severe symptoms promote acute respiratory distress syndrome (ARDS), and ARDS promotes lung infection (Figure 1A). Several stages are involved in the life cycle of SARS-CoV-2. The virus enters the host cell through a cell membrane attachment, and fusions are mediated by the S glycoprotein, replication, transcription, assembly, and release. Two primary processes are involved in the virus entry: (I) receptor binding domain (RBD) and (II) proteolytic activity (transmembrane bound proteases serine 2 and cathepsin L). The SARS-CoV-2 surface contains an S protein that binds to the receptor [14], and it can be cleaved by a furin-like protease into two functional subunits, such as S1 and S2.
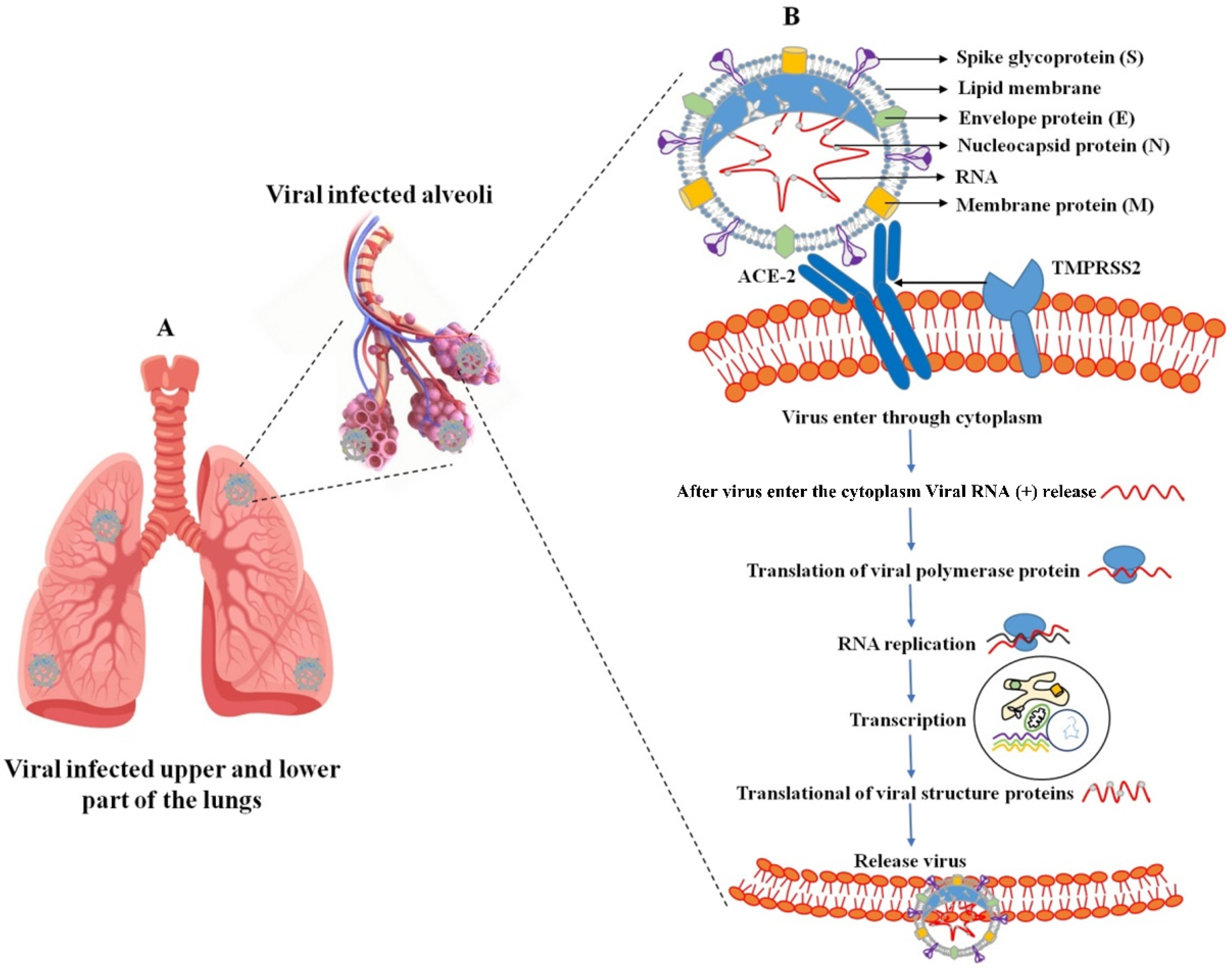
Figure 1. SARS-CoV-2 infection in the upper and lower part of the lungs (A); virus-infected host cell and virus propagation cycle (B). The SARS-CoV-2 virus consists of membrane proteins (M) in the lipid membrane along with envelope proteins (E) and spike glycoproteins linked to the envelope. RNA bound to the nucleocapsid protein exists inside the virus. External binding protein S can enter the lungs by binding to angiotensin-converting enzyme-2 (ACE-2) receptors present in human lung cells. In this case, the virus releases RNA together with genetic information and performs RNA replication to synthesize all components of the virus. Synthetic components are assembled and released out of lung cells.
The receptor binding domain (RBD) of the S1 subunit contains the angiotensin-converting enzyme-2 (ACE2) responsible for binding to the virus receptor, and S2 mainly contains the HR domain. This involves virus fusion and is also divided into HR1 and HR2 [15]. Various types of protease involve the cleavage of proteins at the S1 or S2 sites on the host cell surface by the host transmembrane-bound protease serine 2 (TMPRSS2 early pathway) and utilize the endosomal cathepsin L (CatL) to enter using the late pathway [16][17]. These processes assess membrane fusion between the virus and host cell and also release the viral genome into the cytoplasm [17]. The virus enters the cytoplasm; the viral genomic RNA (sgRNA) is translated into the two polyproteins 1a and 1b (pp1a (ORF1a producing) and pp1b (ORF1b producing)). These proteolytic activities assess the 15–16 nonstructural proteins (NSPs), and each protein has its own specific function. Among them, NSP12 (RNA-dependent RNA polymerase (RdRp)) assembles with several NSPs to form a viral genome replication and transcription complex (RTC). The NSPs 3, 4, and 6 prompted cellular membranes to form double-membrane vesicles (DMVs) [17][18][19]. Transcription of a 5′-set of nested negative-sense sub-genomic RNA molecules (sgRNAs) is used to synthesize the 3′-set of nested positive-sense sgRNAs [20][21]. The structural proteins (S, HE, M, and E) and other related accessory proteins are transformed into the endoplasmic reticulum (ER), but the N protein is translated by cytosolic ribosomes. The assembled virus particles take place in the ER-Golgi intermediate compartment (ERGIC), and new virions are released through exocytosis (Figure 1B). The main functions of a structural protein are S protein (virus entry and fusion), M and E proteins (assembly and production of the virion), and N protein (binds with viral genome and virus release).
At this point, there are no specific antiviral drugs, and before developing the vaccine, people must know how to infect the host cells and what is happening inside the cell. Coronavirus infection promotes lung damage, and those inflammations increase the cytokines and chemokines produced in vivo leading to infection of the alveolar epithelial cells and capillary endothelial cell damage (Figure 2) [22]. After viruses enter a cell, the adaptive immune system responds against an antigen, and B and T cells are involved in this process. The infection cytokines produced by interleukin (IL-6, IL-8, IL-1β), tumor necrosis factor-alpha (TNF-α), and interferon-gamma (IFN-γ) are all accompanied by a severe infection [23]. The chemokines produced were chemokine ligand 3 or macrophage inflammatory protein 1 alpha and 1 beta (MIP1α and 1β), C-C motif chemokine ligand 2 (CCL2), chemokine ligand 5 and 20 (CCL5, CCL20), C-X-C motif chemokine ligand (CXCL1, CXCL2, CXCL8, CXCL10, and CXCL17) [24]. Interleukin-6 (IL-6) is responsible for the innate and acquired immune response and also several other activities during the improvement of acquired immunity against infections, while also responsible for antibody fabrication of B cells and differentiation of dendritic cell and T cell regulations. TNF-α is responsible for cytokines for hyperinflammation through viral infections and cytokines secreted by macrophages, Th17 cells, CD8+ T cells, and DCs. C-reactive protein (CRP) is the prototype of human acute phase proteins (APPs). CRP plays an important role in innate immunity as an early defense mechanism against infections. The elevated levels of IL-6, TNF-α, and C-reaction protein (CRP) have caused more lungs damages. There is currently no treatment for long-term symptoms caused by a SARS-CoV-2 infection. This could be the next major issue to address. Veronese et al. (2022) found olfactory dysfunction from long COVID-19; they used sniff sticks to reduce COVID-19 symptoms, which were excluded by olfactory training/stimulation for up to 30 days compared with the control [25].
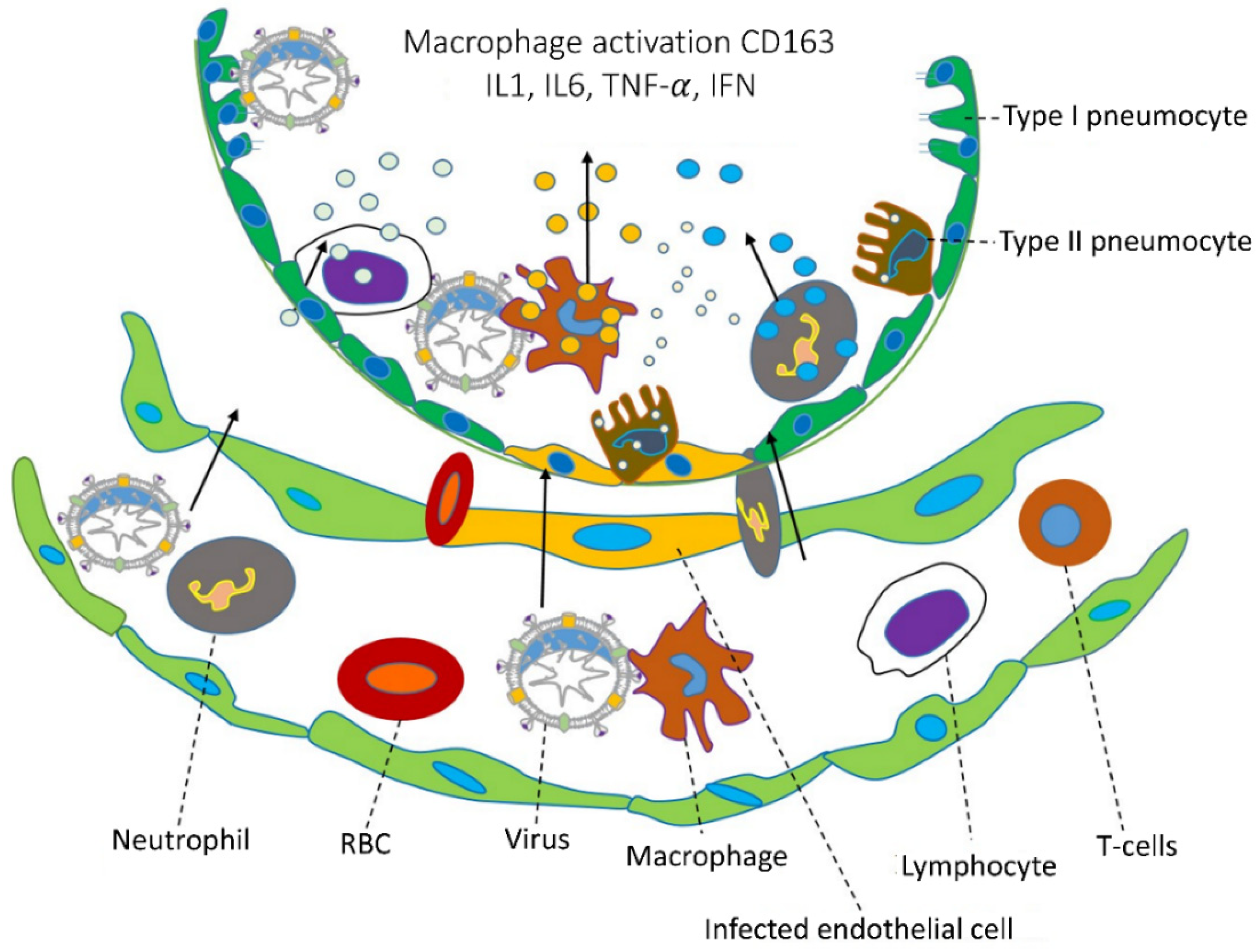
Figure 2. Coronavirus infections and immune approach. During coronavirus infections, the lower part of the lungs collects fluid in the bronchioles and disrupts surface coating types I and II pneumocytes. Additionally, they disrupt the immune response and increase cytokine release, which leads to the accumulation of reactive oxygen species. Various cytokines are secreted by CD163 macrophages activated by a viral infection in infected endothelial cells, and immune responses are caused by IL1, IL6, TNF-α, IFN, etc. Chemical catalysts by this cytokine attract various immune cells, such as neutrophils, lymphocytes, and T cells, through blood vessels. (The specific cells are indicated by dotted lines, and the direction of cell movement is indicated by an arrow).
References
- Hamre, D.; Procknow, J.J. A New Virus Isolated from the Human Respiratory Tract. Proc. Soc. Exp. Biol. Med. 1966, 121, 190–193.
- McIntosh, K.; Dees, J.H.; Becker, W.B.; Kapikian, A.Z.; Chanock, R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA 1967, 57, 933–940.
- van der Hoek, L.; Pyrc, K.; Berkhout, B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006, 30, 760–773.
- Woo, P.C.; Lau, S.K.; Chu, C.M.; Chan, K.H.; Tsoi, H.W.; Huang, Y.; Wong, B.H.; Poon, R.W.; Cai, J.J.; Luk, W.K.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895.
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820.
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266.
- Ye, Z.-W.; Yuan, S.; Yuen, K.-S.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697.
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Chapter Eight—Hosts and Sources of Endemic Human Coronaviruses. In Advances in Virus Research; Kielian, M., Mettenleiter, T.C., Roossinck, M.J.B.T.-A., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 100, pp. 163–188. ISBN 0065-3527.
- Brian, D.A.; Baric, R.S. Coronavirus Genome Structure and Replication BT—Coronavirus Replication and Reverse Genetics; Enjuanes, L., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–30. ISBN 978-3-540-26765-2.
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236.
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48.
- Al-Qaaneh, A.M.; Alshammari, T.; Aldahhan, R.; Aldossary, H.; Alkhalifah, Z.A.; Borgio, J.F. Genome composition and genetic characterization of SARS-CoV-2. Saudi J. Biol. Sci. 2021, 28, 1978–1989.
- Nelson, C.W.; Ardern, Z.; Goldberg, T.L.; Meng, C.; Kuo, C.-H.; Ludwig, C.; Kolokotronis, S.-O.; Wei, X. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. Elife 2020, 9, e59633.
- Renhong, Y.; Yuanyuan, Z.; Yaning, L.; Lu, X.; Yingying, G.; Qiang, Z. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448.
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192.
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8.
- Maier, H.J.; Hawes, P.C.; Cottam, E.M.; Mantell, J.; Verkade, P.; Monaghan, P.; Wileman, T.; Britton, P. Infectious Bronchitis Virus Generates Spherules from Zippered Endoplasmic Reticulum Membranes. MBio 2022, 4, e00801-13.
- Angelini, M.M.; Akhlaghpour, M.; Neuman, B.W.; Buchmeier, M.J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. MBio 2022, 4, e00524-13.
- Masters, P.S. The Molecular Biology of Coronaviruses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2006; Volume 66, pp. 193–292. ISBN 0065-3527.
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263.
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362.
- Li, X.; Geng, M.; Peng, Y.; Meng, L.; Lu, S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020, 10, 102–108.
- Wu, M.-L.; Liu, F.-L.; Sun, J.; Li, X.; He, X.-Y.; Zheng, H.-Y.; Zhou, Y.-H.; Yan, Q.; Chen, L.; Yu, G.-Y.; et al. SARS-CoV-2-triggered mast cell rapid degranulation induces alveolar epithelial inflammation and lung injury. Signal Transduct. Target. Ther. 2021, 6, 428.
- Veronese, N.; Bonica, R.; Cotugno, S.; Tulone, O.; Camporeale, M.; Smith, L.; Trott, M.; Bruyere, O.; Mirarchi, L.; Rizzo, G.; et al. Interventions for Improving Long COVID-19 Symptomatology: A Systematic Review. Viruses 2022, 14, 1863.
More
Information
Subjects:
Virology; Infectious Diseases
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
982
Entry Collection:
COVID-19
Revisions:
3 times
(View History)
Update Date:
25 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




