
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kevina Sonawala | -- | 4142 | 2022-11-18 16:20:22 | | | |
| 2 | Amina Yu | + 10 word(s) | 4152 | 2022-11-21 03:08:39 | | |
Video Upload Options
Initially entitled as junk matter, non-coding RNAs are an exceptional class of RNAs constituting a majority of the transcriptional output in living cells, which are not translated into functional proteins. They are not only responsible for regulating the expression of the gene at the transcriptional and post-transcriptional stages but also for mediating various cellular processes such as heterochromatin formation, epigenetic modifications, signal transduction and so on. It is quite evident from one research that the abnormal expression of LncRNAs plays a significant role in cancer stem cells (CSCs)’ metabolism. hey regulate gene expression by the following approaches: as a modulator of gene expression; as a decoy to lead the transcription factor elsewhere from a target site; as a competitor to hinder the attachment of other molecules to the target site; as a chaperone for molecules to attach to a certain segment and as a scaffold that enhances the association of different proteins into different complexes.
1. Focus on LncRNAs
2. The Interplay of LncRNAs in CSCs Signaling Pathways
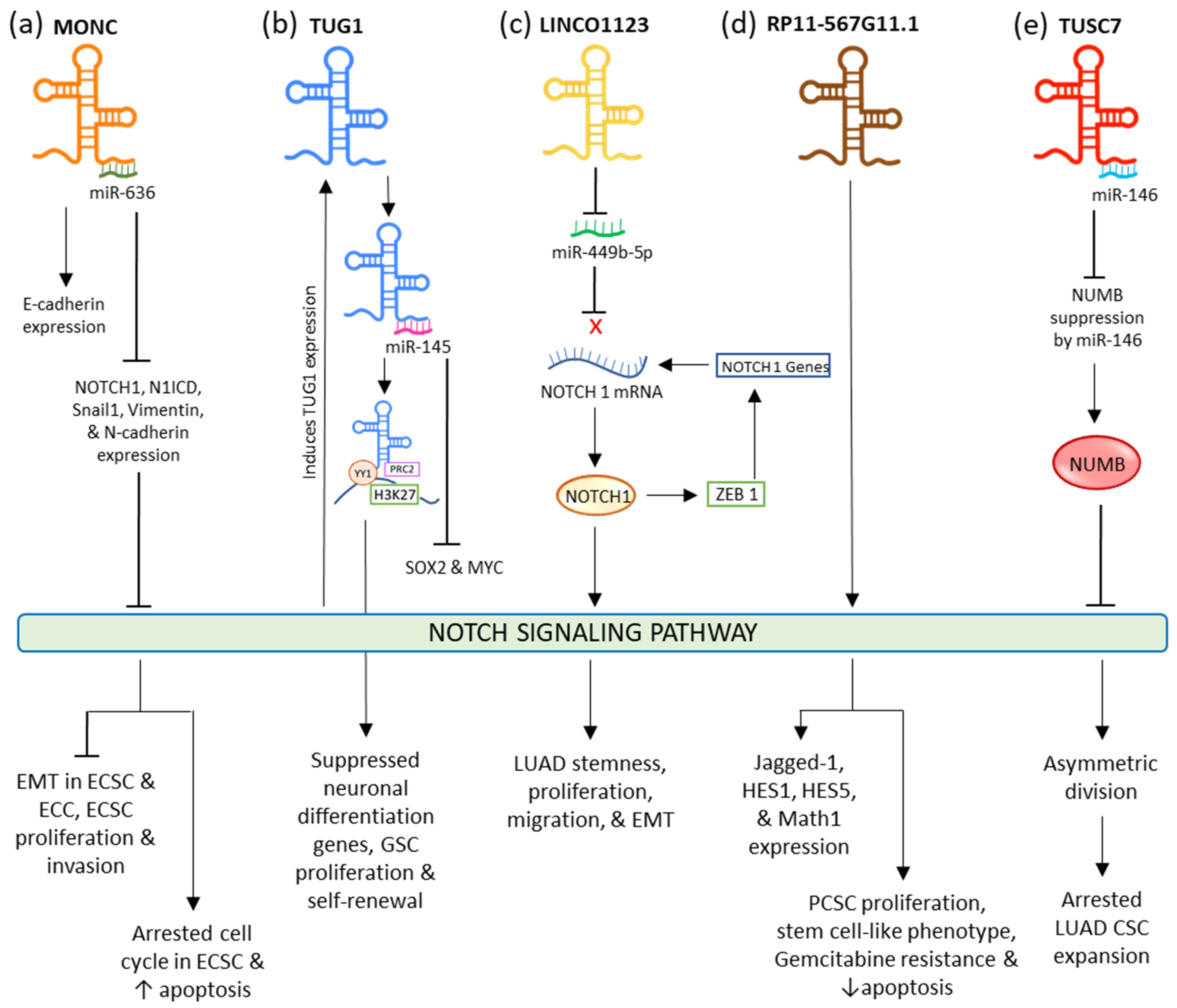
2.1. NOTCH Signaling Pathway
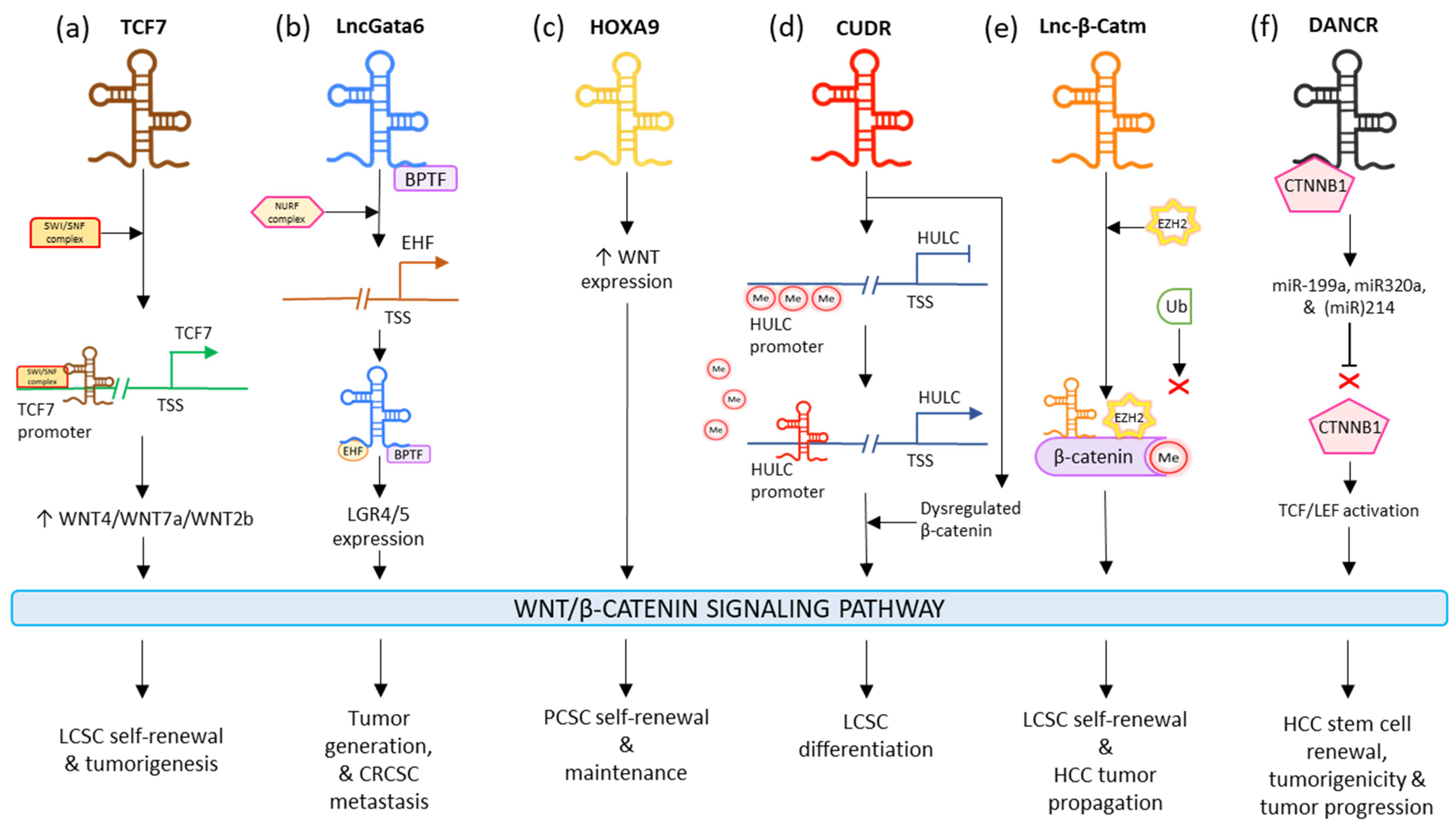
2.2. WNT Signaling Pathway
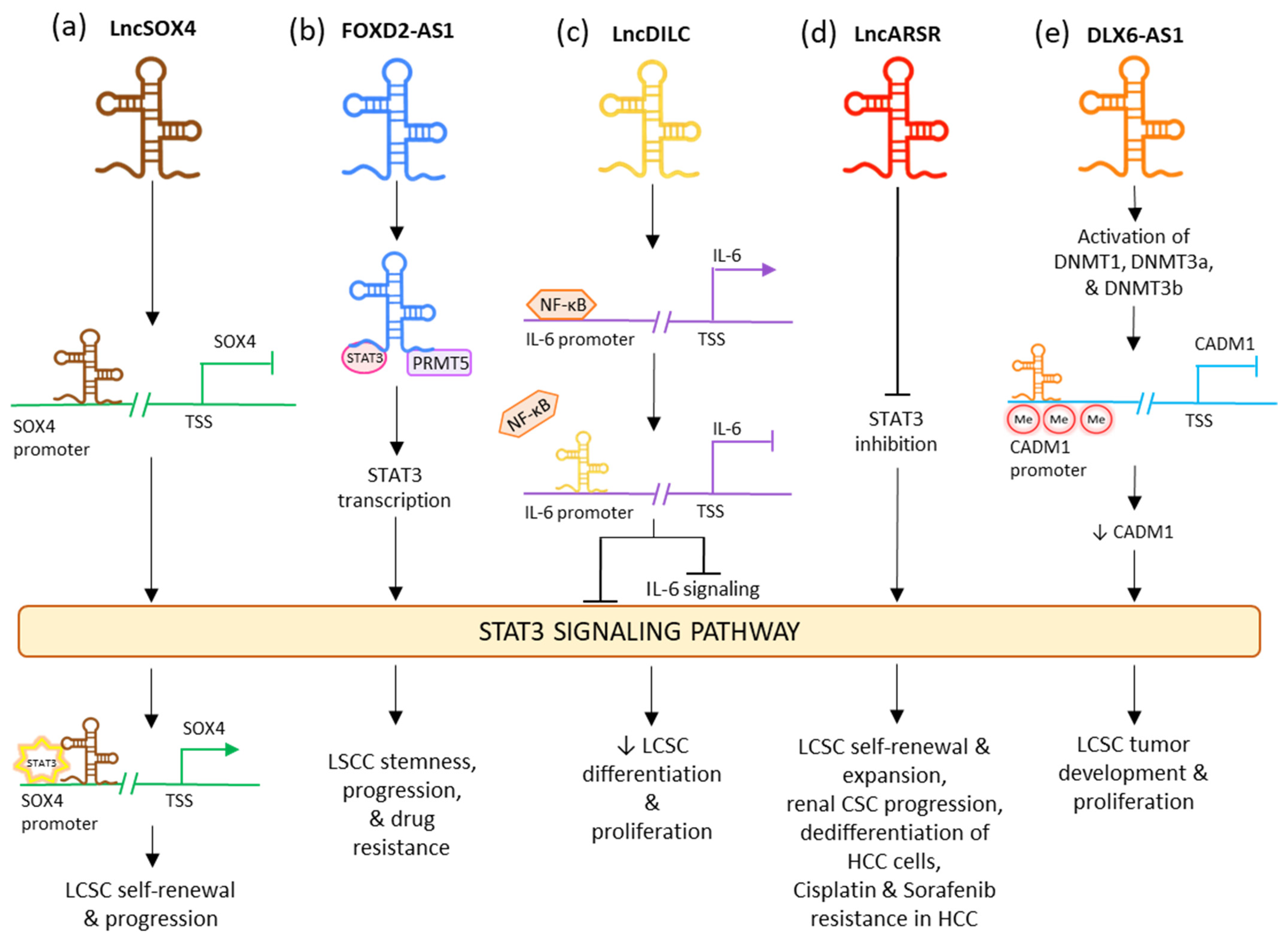
2.3. STAT Signaling Pathway
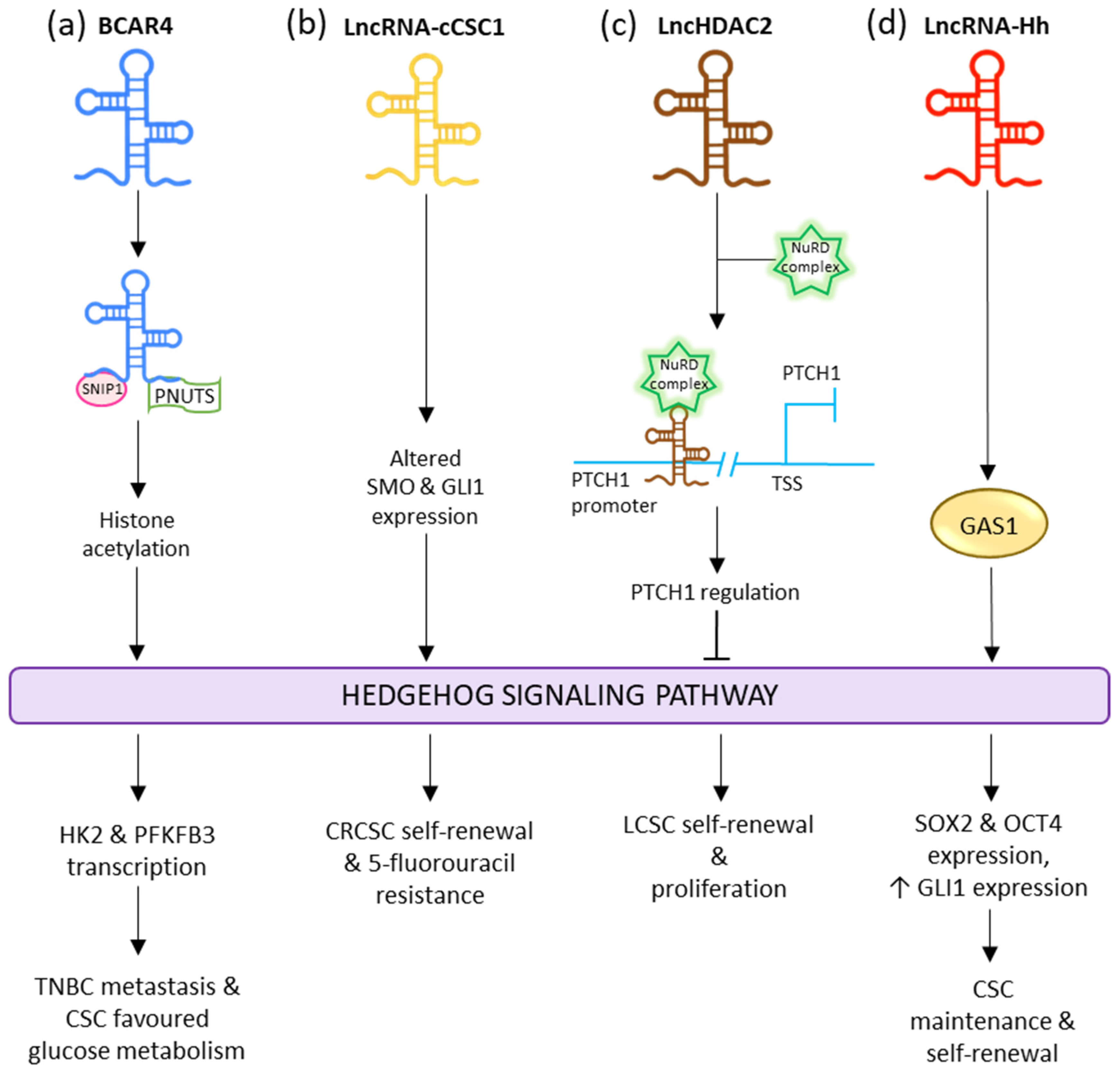
2.4. Hedgehog Signaling Pathway
References
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914.
- Li, W.; Jiang, P.; Sun, X.; Xu, S.; Ma, X.; Zhan, R. Suppressing H19 Modulates Tumorigenicity and Stemness in U251 and U87MG Glioma Cells. Cell. Mol. Neurobiol. 2016, 36, 1219–1227.
- Peng, F.; Li, T.T.; Wang, K.L.; Xiao, G.Q.; Wang, J.H.; Zhao, H.D.; Kang, Z.J.; Fan, W.J.; Zhu, L.L.; Li, M.; et al. H19/Let-7/LIN28 Reciprocal Negative Regulatory Circuit Promotes Breast Cancer Stem Cell Maintenance. Cell Death Dis. 2017, 8, e2569.
- Feng, S.; Yao, J.; Chen, Y.; Geng, P.; Zhang, H.; Ma, X.; Zhao, J.; Yu, X. Expression and Functional Role of Reprogramming-Related Long Noncoding RNA (LincRNA-ROR) in Glioma. J. Mol. Neurosci. 2015, 56, 623–630.
- Wu, M.; An, J.; Zheng, Q.; Xin, X.; Lin, Z.; Li, X.; Li, H.; Lu, D. Double Mutant P53 (N340Q/L344R) Promotes Hepatocarcinogenesis through Upregulation of Pim1 Mediated by PKM2 and LncRNA CUDR. Oncotarget 2016, 7, 66525–66539.
- Pu, H.; Zheng, Q.; Li, H.; Wu, M.; An, J.; Gui, X.; Li, T.; Lu, D. CUDR Promotes Liver Cancer Stem Cell Growth through Upregulating TERT and C-Myc. Oncotarget 2015, 6, 40775–40798.
- Li, L.; Dang, Q.; Xie, H.; Yang, Z.; He, D.; Liang, L.; Song, W.; Yeh, S.; Chang, C. Correction: Infiltrating Mast Cells Enhance Prostate Cancer Invasion via Altering LncRNA-HOTAIR/PRC2-Androgen Receptor (AR)-MMP9 Signals and Increased Stem/Progenitor Cell Population. Oncotarget 2016, 7, 83828.
- Alves, C.P.; Fonseca, A.S.; Muys, B.R.; Bueno, R.D.B.E.L.; Burger, M.C.; De Souza, J.E.S.; Valente, V.; Zago, M.A.; Silva, W.A. Brief Report: The LincRNA Hotair Is Required for Epithelial-to-Mesenchymal Transition and Stemness Maintenance of Cancer Cell Lines. Stem Cells 2013, 31, 2827–2832.
- Galasso, M.; Dama, P.; Previati, M.; Sandhu, S.; Palatini, J.; Coppola, V.; Warner, S.; Sana, M.E.; Zanella, R.; Abujarour, R.; et al. A Large Scale Expression Study Associates Uc.283-plus LncRNA with Pluripotent Stem Cells and Human Glioma. Genome Med. 2014, 6, 76.
- Wang, Y.; Wang, Y.; Li, J.; Zhang, Y.; Yin, H.; Han, B. CRNDE, a Long-Noncoding RNA, Promotes Glioma Cell Growth and Invasion through MTOR Signaling. Cancer Lett. 2015, 367, 122–128.
- Ghafouri-Fard, S.; Dashti, S.; Farsi, M.; Taheri, M.; Mousavinejad, S.A. X-Inactive-Specific Transcript: Review of Its Functions in the Carcinogenesis. Front. Cell Dev. Biol. 2021, 9, 690522.
- Yang, L.; Lin, C.; Jin, C.; Yang, J.C.; Tanasa, B.; Li, W.; Merkurjev, D.; Ohgi, K.A.; Meng, D.; Zhang, J.; et al. LncRNA-Dependent Mechanisms of Androgen-Receptor-Regulated Gene Activation Programs. Nature 2013, 500, 598–602.
- Popadiuk, C.M.; Xiong, J.; Wells, M.G.; Andrews, P.G.; Dankwa, K.; Hirasawa, K.; Lake, B.B.; Kao, K.R. Antisense Suppression of Pygopus2 Results in Growth Arrest of Epithelial Ovarian Cancer. Clin. Cancer Res. 2006, 12, 2216–2223.
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of P53 by MEG3 Non-Coding RNA. J. Biol. Chem. 2007, 282, 24731–24742.
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The Functional Role of Long Non-Coding RNA in Human Carcinomas. Mol. Cancer 2011, 10, 38.
- Zhang, X.; Gejman, R.; Mahta, A.; Zhong, Y.; Rice, K.A.; Zhou, Y.; Cheunsuchon, P.; Louis, D.N.; Klibanski, A. Maternally Expressed Gene 3, an Imprinted Noncoding RNA Gene, Is Associated with Meningioma Pathogenesis and Progression. Cancer Res. 2010, 70, 2350–2358.
- Benetatos, L.; Hatzimichael, E.; Dasoula, A.; Dranitsaris, G.; Tsiara, S.; Syrrou, M.; Georgiou, I.; Bourantas, K.L. CpG Methylation Analysis of the MEG3 and SNRPN Imprinted Genes in Acute Myeloid Leukemia and Myelodysplastic Syndromes. Leuk. Res. 2010, 34, 148–153.
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. MicroRNA-29 Can Regulate Expression of the Long Non-Coding RNA Gene MEG3 in Hepatocellular Cancer. Oncogene 2011, 30, 4750–4756.
- Chiba, S. Concise Review: Notch Signaling in Stem Cell Systems. Stem Cells 2006, 24, 2437–2447.
- Karamboulas, C.; Ailles, L. Developmental Signaling Pathways in Cancer Stem Cells of Solid Tumors. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 2481–2495.
- Ellisen, L.W.; Bird, J.; West, D.C.; Soreng, A.L.; Reynolds, T.C.; Smith, S.D.; Sklar, J. TAN-1, the Human Homolog of the Drosophila Notch Gene, Is Broken by Chromosomal Translocations in T Lymphoblastic Neoplasms. Cell 1991, 66, 649–661.
- Ma, Y.-C.; Shi, C.; Zhang, Y.-N.; Wang, L.-G.; Liu, H.; Jia, H.-T.; Zhang, Y.-X.; Sarkar, F.H.; Wang, Z.-S. The Tyrosine Kinase C-Src Directly Mediates Growth Factor-Induced Notch-1 and Furin Interaction and Notch-1 Activation in Pancreatic Cancer Cells. PLoS ONE 2012, 7, e33414.
- Wu, F.; Stutzman, A.; Mo, Y.Y. Notch Signaling and Its Role in Breast Cancer. Front. Biosci. 2007, 12, 4370–7383.
- Song, L.L.; Peng, Y.; Yun, J.; Rizzo, P.; Chaturvedi, V.; Weijzen, S.; Kast, W.M.; Stone, P.J.B.; Santos, L.; Loredo, A.; et al. Notch-1 Associates with IKKα and Regulates IKK Activity in Cervical Cancer Cells. Oncogene 2008, 27, 5833–5844.
- Qiao, L.; Wong, B.C.Y. Role of Notch Signaling in Colorectal Cancer. Carcinogenesis 2009, 30, 1979–1986.
- Zhou, W.; Fu, X.-Q.; Zhang, L.-L.; Zhang, J.; Huang, X.; Lu, X.-H.; Shen, L.; Liu, B.-N.; Liu, J.; Luo, H.-S.; et al. The AKT1/NF-KappaB/Notch1/PTEN Axis Has an Important Role in Chemoresistance of Gastric Cancer Cells. Cell Death Dis. 2013, 4, e847.
- Zhang, Y.; Li, B.; Ji, Z.Z.; Zheng, P.S. Notch1 Regulates the Growth of Human Colon Cancers. Cancer 2010, 116, 5207–5218.
- Ranganathan, P.; Weaver, K.L.; Capobianco, A.J. Notch Signalling in Solid Tumours: A Little Bit of Everything but Not All the Time. Nat. Rev. Cancer 2011, 11, 338–351.
- Abel, E.V.; Kim, E.J.; Wu, J.; Hynes, M.; Bednar, F.; Proctor, E.; Wang, L.; Dziubinski, M.L.; Simeone, D.M. The Notch Pathway Is Important in Maintaining the Cancer Stem Cell Population in Pancreatic Cancer. PLoS ONE 2014, 9, e91983.
- Kannan, S.; Sutphin, R.M.; Hall, M.G.; Golfman, L.S.; Fang, W.; Nolo, R.M.; Akers, L.J.; Hammitt, R.A.; McMurray, J.S.; Kornblau, S.M.; et al. Notch Activation Inhibits AML Growth and Survival: A Potential Therapeutic Approach. J. Exp. Med. 2013, 210, 321–337.
- Lefort, K.; Mandinova, A.; Ostano, P.; Kolev, V.; Calpini, V.; Kolfschoten, I.; Devgan, V.; Lieb, J.; Raffoul, W.; Hohl, D.; et al. Notch1 Is a P53 Target Gene Involved in Human Keratinocyte Tumor Suppression through Negative Regulation of ROCK1/2 and MRCKα Kinases. Genes Dev. 2007, 21, 562–577.
- Konishi, J.; Yi, F.; Chen, X.; Vo, H.; Carbone, D.P.; Dang, T.P. Notch3 Cooperates with the EGFR Pathway to Modulate Apoptosis through the Induction of Bim. Oncogene 2009, 29, 589–596.
- Viatour, P.; Ehmer, U.; Saddic, L.A.; Dorrell, C.; Andersen, J.B.; Lin, C.; Zmoos, A.F.; Mazur, P.K.; Schaffer, B.E.; Ostermeier, A.; et al. Notch Signaling Inhibits Hepatocellular Carcinoma Following Inactivation of the RB Pathway. J. Exp. Med. 2011, 208, 1963–1976.
- Gupta, A.; Wang, Y.; Browne, C.; Kim, S.; Case, T.; Paul, M.; Wills, M.L.; Matusik, R.J. Neuroendocrine Differentiation in the 12T-10 Transgenic Prostate Mouse Model Mimics Endocrine Differentiation of Pancreatic Beta Cells. Prostate 2008, 68, 50–60.
- Parr, C.; Watkins, G.; Jiang, W.G. The Possible Correlation of Notch-1 and Notch-2 with Clinical Outcome and Tumour Clinicopathological Parameters in Human Breast Cancer. Int. J. Mol. Med. 2004, 14, 779–786.
- Li, Y.; Huo, J.; He, J.; Ma, X. LncRNA MONC Suppresses the Malignant Phenotype of Endometrial Cancer Stem Cells and Endometrial Carcinoma Cells by Regulating the MiR-636/GLCE Axis. Cancer Cell Int. 2021, 21, 331.
- Tan, J.; Qiu, K.; Li, M.; Liang, Y. Double-Negative Feedback Loop between Long Non-Coding RNA TUG1 and MiR-145 Promotes Epithelial to Mesenchymal Transition and Radioresistance in Human Bladder Cancer Cells. FEBS Lett. 2015, 589, 3175–3181.
- Cao, W.J.; Wu, H.L.; He, B.S.; Zhang, Y.S.; Zhang, Z.Y. Analysis of Long Non-Coding RNA Expression Profiles in Gastric Cancer. World J. Gastroenterol. 2013, 19, 3658–3664.
- Zhang, Q.; Geng, P.L.; Yin, P.; Wang, X.L.; Jia, J.P.; Yao, J. Down-Regulation of Long Non-Coding RNA TUG1 Inhibits Osteosarcoma Cell Proliferation and Promotes Apoptosis. Asian Pac. J. Cancer Prev. 2013, 14, 2311–2315.
- Zhang, E.-B.; Yin, D.-D.; Sun, M.; Kong, R.; Liu, X.-H.; You, L.-H.; Han, L.; Xia, R.; Wang, K.-M.; Yang, J.-S.; et al. P53-Regulated Long Non-Coding RNA TUG1 Affects Cell Proliferation in Human Non-Small Cell Lung Cancer, Partly through Epigenetically Regulating HOXB7 Expression. Cell Death Dis. 2014, 5, e1243.
- Zhang, M.; Han, Y.; Zheng, Y.; Zhang, Y.; Zhao, X.; Gao, Z.; Liu, X. ZEB1-Activated LINC01123 Accelerates the Malignancy in Lung Adenocarcinoma through NOTCH Signaling Pathway. Cell Death Dis. 2020, 11, 981.
- Huang, R.; Nie, W.; Yao, K.; Chou, J. Depletion of the LncRNA RP11-567G11.1 Inhibits Pancreatic Cancer Progression. Biomed. Pharmacother. 2019, 112, 108685.
- Huang, G.; Wang, M.; Li, X.; Wu, J.; Chen, S.; Du, N.; Li, K.; Wang, J.; Xu, C.; Ren, H.; et al. TUSC7 Suppression of Notch Activation through Sponging MiR-146 Recapitulated the Asymmetric Cell Division in Lung Adenocarcinoma Stem Cells. Life Sci. 2019, 232, 116630.
- Acebron, S.P.; Karaulanov, E.; Berger, B.S.; Huang, Y.L.; Niehrs, C. Mitotic Wnt Signaling Promotes Protein Stabilization and Regulates Cell Size. Mol. Cell 2014, 54, 663–674.
- Atlasi, Y.; Noori, R.; Gaspar, C.; Franken, P.; Sacchetti, A.; Rafati, H.; Mahmoudi, T.; Decraene, C.; Calin, G.A.; Merrill, B.J.; et al. Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation. PLoS Genet. 2013, 9, e1003424.
- Clevers, H.; Loh, K.M.; Nusse, R. An Integral Program for Tissue Renewal and Regeneration: Wnt Signaling and Stem Cell Control. Science 2014, 3, 346.
- Green, J.L.; Inoue, T.; Sternberg, P.W. Opposing Wnt Pathways Orient Cell Polarity during Organogenesis. Cell 2008, 134, 646–656.
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt Signaling in Cancer. Oncogene 2017, 36, 1461–1473.
- Grumolato, L.; Liu, G.; Mong, P.; Mudbhary, R.; Biswas, R.; Arroyave, R.; Vijayakumar, S.; Economides, A.N.; Aaronson, S.A. Canonical and Noncanonical Wnts Use a Common Mechanism to Activate Completely Unrelated Coreceptors. Genes Dev. 2010, 24, 2517–2530.
- Katoh, M. Canonical and Non-Canonical WNT Signaling in Cancer Stem Cells and Their Niches: Cellular Heterogeneity, Omics Reprogramming, Targeted Therapy and Tumor Plasticity (Review). Int. J. Oncol. 2017, 51, 1357–1369.
- Dieter, S.M.; Glimm, H.; Ball, C.R. Colorectal Cancer-initiating Cells Caught in the Act. EMBO Mol. Med. 2017, 9, 856–858.
- Kahn, M. Can We Safely Target the WNT Pathway? Nat. Rev. Drug Discov. 2014, 13, 513–532.
- Mirabelli, C.K.; Nusse, R.; Tuveson, D.A.; Williams, B.O. Perspectives on the Role of Wnt Biology in Cancer. Sci. Signal. 2019, 12, eaay4494.
- Amin, N.; Cavallaro, U. The Wnt Signaling Pathways and Cell Adhesion. Front. Biosci. 2012, 17, 784–804.
- Schatoff, E.M.; Leach, B.I.; Dow, L.E. WNT Signaling and Colorectal Cancer. Curr. Color. Cancer Rep. 2017, 13, 101–110.
- Klarmann, G.J.; Decker, A.; Farrar, W.L. Epigenetic Gene Silencing in the Wnt Pathway in Breast Cancer. Epigenetics 2008, 3, 59–63.
- Wang, Y.; He, L.; Du, Y.; Zhu, P.; Huang, G.; Luo, J.; Yan, X.; Ye, B.; Li, C.; Xia, P.; et al. The Long Noncoding RNA LncTCF7 Promotes Self-Renewal of Human Liver Cancer Stem Cells through Activation of Wnt Signaling. Cell Stem Cell 2015, 16, 413–425.
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Cell Stem Cell 2014, 14, 342–356.
- Malanchi, I.; Santamaria-Martínez, A.; Susanto, E.; Peng, H.; Lehr, H.A.; Delaloye, J.F.; Huelsken, J. Interactions between Cancer Stem Cells and Their Niche Govern Metastatic Colonization. Nature 2012, 481, 85–91.
- Zhu, P.; Wu, J.; Wang, Y.; Zhu, X.; Lu, T.; Liu, B.; He, L.; Ye, B.; Wang, S.; Meng, S.; et al. LncGata6 Maintains Stemness of Intestinal Stem Cells and Promotes Intestinal Tumorigenesis. Nat. Cell Biol. 2018, 20, 1134–1144.
- Li, Z.; Zhao, L.; Wang, Q. Overexpression of Long Non-Coding RNA HOTTIP Increases Chemoresistance of Osteosarcoma Cell by Activating the Wnt/β-Catenin Pathway. Am. J. Transl. Res. 2016, 8, 2385.
- Gui, X.; Li, H.; Li, T.; Pu, H.; Lu, D. Long Noncoding RNA CUDR Regulates HULC and β-Catenin to Govern Human Liver Stem Cell Malignant Differentiation. Mol. Ther. 2015, 23, 1843–1853.
- Wang, J.; Lei, Z.J.; Guo, Y.; Wang, T.; Qin, Z.Y.; Xiao, H.L.; Fan, L.L.; Chen, D.F.; Bian, X.W.; Liu, J.; et al. MiRNA-Regulated Delivery of LincRNA-P21 Suppresses β-Catenin Signaling and Tumorigenicity of Colorectal Cancer Stem Cells. Oncotarget 2015, 6, 37852–37870.
- Luo, M.; Li, Z.; Wang, W.; Zeng, Y.; Liu, Z.; Qiu, J. Long Non-Coding RNA H19 Increases Bladder Cancer Metastasis by Associating with EZH2 and Inhibiting E-Cadherin Expression. Cancer Lett. 2013, 333, 213–221.
- Zhu, P.; Wang, Y.; Huang, G.; Ye, B.; Liu, B.; Wu, J.; Du, Y.; He, L.; Fan, Z. Lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016, 23, 631–639.
- Yuan, S.-X.; Wang, J.; Yang, F.; Tao, Q.-F.; Zhang, J.; Wang, L.-L.; Yang, Y.; Liu, H.; Wang, Z.-G.; Xu, Q.-G.; et al. Long Noncoding RNA DANCR Increases Stemness Features of Hepatocellular Carcinoma by Derepression of CTNNB1. Hepatology 2016, 63, 499–511.
- Yan, J.; Dang, Y.; Liu, S.; Zhang, Y.; Zhang, G. LncRNA HOTAIR Promotes Cisplatin Resistance in Gastric Cancer by Targeting MiR-126 to Activate the PI3K/AKT/MRP1 Genes. Tumor Biol. 2016, 37, 16345–16355.
- Gatta, L.B.; Melocchi, L.; Bugatti, M.; Missale, F.; Lonardi, S.; Zanetti, B.; Cristinelli, L.; Belotti, S.; Simeone, C.; Ronca, R.; et al. Hyper-Activation of STAT3 Sustains Progression of Non-Papillary Basal-Type Bladder Cancer via FOSL1 Regulome. Cancers 2019, 11, 1219.
- Kim, J.H.; Choi, H.S.; Kim, S.L.; Lee, D.S. The PAK1-Stat3 Signaling Pathway Activates IL-6 Gene Transcription and Human Breast Cancer Stem Cell Formation. Cancers 2019, 11, 1527.
- White, C.L.; Jayasekara, W.S.N.; Picard, D.; Chen, J.; Watkins, D.N.; Cain, J.E.; Remke, M.; Gough, D.J. A Sexually Dimorphic Role for STAT3 in Sonic Hedgehog Medulloblastoma. Cancers 2019, 11, 1702.
- Yun, J.W.; Lee, S.; Kim, H.M.; Chun, S.; Engleman, E.G.; Kim, H.C.; Kang, E.S. A Novel Type of Blood Biomarker: Distinct Changes of Cytokine-Induced Stat Phosphorylation in Blood t Cells between Colorectal Cancer Patients and Healthy Individuals. Cancers 2019, 11, 1157.
- Severin, F.; Frezzato, F.; Visentin, A.; Martini, V.; Trimarco, V.; Carraro, S.; Tibaldi, E.; Maria Brunati, A.; Piazza, F.; Semenzato, G.; et al. In Chronic Lymphocytic Leukemia the JAK2/STAT3 Pathway Is Constitutively Activated and Its Inhibition Leads to CLL Cell Death Unaffected by the Protective Bone Marrow Microenvironment. Cancers 2019, 11, 1939.
- Morgan, E.L.; Macdonald, A. JAK2 Inhibition Impairs Proliferation and Sensitises Cervical Cancer Cells to Cisplatin-Induced Cell Death. Cancers 2019, 11, 1934.
- Basu, R.; Kulkarni, P.; Qian, Y.; Walsh, C.; Arora, P.; Davis, E.; Duran-Ortiz, S.; Funk, K.; Ibarra, D.; Kruse, C.; et al. Growth Hormone Upregulates Melanocyte-Inducing Transcription Factor Expression and Activity via JAK2-STAT5 and SRC Signaling in GH Receptor-Positive Human Melanoma. Cancers 2019, 11, 1352.
- Maurer, B.; Kollmann, S.; Pickem, J.; Hoelbl-Kovacic, A.; Sexl, V. STAT5A and STAT5B—Twins with Different Personalities in Hematopoiesis and Leukemia. Cancers 2019, 11, 1726.
- Moll, H.P.; Mohrherr, J.; Blaas, L.; Musteanu, M.; Stiedl, P.; Grabner, B.; Zboray, K.; König, M.; Stoiber, D.; Rülicke, T.; et al. A Mouse Model to Assess STAT3 and STAT5A/B Combined Inhibition in Health and Disease Conditions. Cancers 2019, 11, 1226.
- Valle-Mendiola, A.; Soto-Cruz, I. Energy Metabolism in Cancer: The Roles of STAT3 and STAT5 in the Regulation of Metabolism-Related Genes. Cancers 2020, 12, 124.
- Seif, F.; Khoshmirsafa, M.; Aazami, H.; Mohsenzadegan, M.; Sedighi, G.; Bahar, M. The Role of JAK-STAT Signaling Pathway and Its Regulators in the Fate of T Helper Cells. Cell Commun. Signal. 2017, 15, 23.
- Birnie, R.; Bryce, S.D.; Roome, C.; Dussupt, V.; Droop, A.; Lang, S.H.; Berry, P.A.; Hyde, C.F.; Lewis, J.L.; Stower, M.J.; et al. Gene Expression Profiling of Human Prostate Cancer Stem Cells Reveals a Pro-Inflammatory Phenotype and the Importance of Extracellular Matrix Interactions. Genome Biol. 2008, 9, R83.
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J.; Margolick, J.B.; Liotta, L.A.; Petricoin, E.; Zhang, Y. Activation of the PTEN/MTOR/STAT3 Pathway in Breast Cancer Stem-like Cells Is Required for Viability and Maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163.
- Sherry, M.M.; Reeves, A.; Wu, J.K.; Cochran, B.H. STAT3 Is Required for Proliferation and Maintenance of Multipotency in Glioblastoma Stem Cells. Stem Cells 2009, 27, 2383–2392.
- Cook, A.M.; Li, L.; Ho, Y.; Lin, A.; Li, L.; Stein, A.; Forman, S.; Perrotti, D.; Jove, R.; Bhatia, R. Role of Altered Growth Factor Receptor-Mediated JAK2 Signaling in Growth and Maintenance of Human Acute Myeloid Leukemia Stem Cells. Blood 2014, 123, 2826–2837.
- Zhang, H.F.; Lai, R. STAT3 in Cancer-Friend or Foe? Cancers 2014, 6, 1408–1440.
- Vultur, A.; Cao, J.; Arulanandam, R.; Turkson, J.; Jove, R.; Greer, P.; Craig, A.; Elliott, B.; Raptis, L. Cell-to-Cell Adhesion Modulates Stat3 Activity in Normal and Breast Carcinoma Cells. Oncogene 2004, 23, 2600–2616.
- Steinman, R.A.; Wentzel, A.; Lu, Y.; Stehle, C.; Grandis, J.R. Activation of Stat3 by Cell Confluence Reveals Negative Regulation of Stat3 by Cdk2. Oncogene 2003, 22, 3608–3615.
- Gkouveris, I.; Nikitakis, N.; Karanikou, M.; Rassidakis, G.; Sklavounou, A. Erk1/2 Activation and Modulation of STAT3 Signaling in Oral Cancer. Oncol. Rep. 2014, 32, 2175–2182.
- Yan, S.; Li, Z.; Thiele, C.J.; Yan, S.; Li, Z.; Thiele, C.J. Inhibition of STAT3 with Orally Active JAK Inhibitor, AZD1480, Decreases Tumor Growth in Neuroblastoma and Pediatric Sarcomas In Vitro and In Vivo. Oncotarget 2013, 4, 433–445.
- Ivanov, V.N.; Bhoumik, A.; Krasilnikov, M.; Raz, R.; Owen-Schaub, L.B.; Levy, D.; Horvath, C.M.; Ronai, Z. Cooperation between STAT3 and C-Jun Suppresses Fas Transcription. Mol. Cell 2001, 7, 517–528.
- Barré, B.; Avril, S.; Coqueret, O. Opposite Regulation of Myc and P21 Waf1 Transcription by STAT3 Proteins. J. Biol. Chem. 2003, 278, 2990–2996.
- Kroon, P.; Berry, P.A.; Stower, M.J.; Rodrigues, G.; Mann, V.M.; Simms, M.; Bhasin, D.; Chettiar, S.; Li, C.; Li, P.K.; et al. JAK-STAT Blockade Inhibits Tumor Initiation and Clonogenic Recovery of Prostate Cancer Stem-like Cells. Cancer Res. 2013, 73, 5288–5298.
- Qu, Y.; Oyan, A.M.; Liu, R.; Hua, Y.; Zhang, J.; Hovland, R.; Popa, M.; Liu, X.; Brokstad, K.A.; Simon, R.; et al. Generation of Prostate Tumor–Initiating Cells Is Associated with Elevation of Reactive Oxygen Species and IL-6/STAT3 Signaling. Cancer Res. 2013, 73, 7090–7100.
- Rybak, A.P.; Bristow, R.G.; Kapoor, A. Prostate Cancer Stem Cells: Deciphering the Origins and Pathways Involved in Prostate Tumorigenesis and Aggression. Oncotarget 2015, 6, 1900–1919.
- Zhao, D.; Pan, C.; Sun, J.; Gilbert, C.; Drews-Elger, K.; Azzam, D.J.; Picon-Ruiz, M.; Kim, M.; Ullmer, W.; El-Ashry, D.; et al. VEGF Drives Cancer-Initiating Stem Cells through VEGFR-2/Stat3 Signaling to Upregulate Myc and SOX2. Oncogene 2014, 34, 3107–3119.
- Gu, L.Q.; Xing, X.L.; Cai, H.; Si, A.F.; Hu, X.R.; Ma, Q.Y.; Zheng, M.L.; Wang, R.Y.; Li, H.Y.; Zhang, X.P. Long Non-Coding RNA DILC Suppresses Cell Proliferation and Metastasis in Colorectal Cancer. Gene 2018, 666, 18–26.
- Wang, X.; Sun, W.; Shen, W.; Xia, M.; Chen, C.; Xiang, D.; Ning, B.; Cui, X.; Li, H.; Li, X.; et al. Long Non-Coding RNA DILC Regulates Liver Cancer Stem Cells via IL-6/STAT3 Axis. J. Hepatol. 2016, 64, 1283–1294.
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An Epigenetic Switch Involving NF-ΚB, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell 2009, 139, 693–706.
- Kagoya, Y.; Yoshimi, A.; Kataoka, K.; Nakagawa, M.; Kumano, K.; Arai, S.; Kobayashi, H.; Saito, T.; Iwakura, Y.; Kurokawa, M. Positive Feedback between NF-ΚB and TNF-α Promotes Leukemia-Initiating Cell Capacity. J. Clin. Investig. 2014, 124, 528–542.
- Magagula, L.; Gagliardi, M.; Naidoo, J.; Mhlanga, M. Lnc-Ing Inflammation to Disease. Biochem. Soc. Trans. 2017, 45, 953–962.
- Chen, Z.; Huang, L.; Wu, Y.; Zhai, W.; Zhu, P.; Gao, Y. LncSOX4 Promotes the Self-Renewal of Liver Tumour-Initiating Cells through Stat3-Mediated SOX4 Expression. Nat. Commun. 2016, 7, 12598.
- Li, W.; Chen, Y.; Nie, X. Regulatory Mechanisms of LncRNAs and Their Target Gene Signaling Pathways in Laryngeal Squamous Cell Carcinoma. Front. Pharmacol. 2020, 11, 1140.
- Yang, C.; Cai, W.C.; Dong, Z.T.; Guo, J.W.; Zhao, Y.J.; Sui, C.J.; Yang, J. mei LncARSR Promotes Liver Cancer Stem Cells Expansion via STAT3 Pathway. Gene 2019, 687, 73–81.
- Wu, D.M.; Zheng, Z.H.; Zhang, Y.B.; Fan, S.H.; Zhang, Z.F.; Wang, Y.J.; Zheng, Y.L.; Lu, J. Down-Regulated LncRNA DLX6-AS1 Inhibits Tumorigenesis through STAT3 Signaling Pathway by Suppressing CADM1 Promoter Methylation in Liver Cancer Stem Cells. J. Exp. Clin. Cancer Res. 2019, 38, 237.
- Yang, L.; Xie, G.; Fan, Q.; Xie, J. Activation of the Hedgehog-Signaling Pathway in Human Cancer and the Clinical Implications. Oncogene 2010, 29, 469–481.
- Peacock, C.D.; Wang, Q.; Gesell, G.S.; Corcoran-Schwartz, I.M.; Jones, E.; Kim, J.; Devereux, W.L.; Rhodes, J.T.; Huff, C.A.; Beachy, P.A.; et al. Hedgehog Signaling Maintains a Tumor Stem Cell Compartment in Multiple Myeloma. Proc. Natl. Acad. Sci. USA 2007, 104, 4048–4053.
- Dembinski, J.L.; Krauss, S. Characterization and Functional Analysis of a Slow Cycling Stem Cell-like Subpopulation in Pancreas Adenocarcinoma. Clin. Exp. Metastasis 2009, 26, 611–623.
- Liu, S.; Dontu, G.; Mantle, I.D.; Patel, S.; Ahn, N.S.; Jackson, K.W.; Suri, P.; Wicha, M.S. Hedgehog Signaling and Bmi-1 Regulate Self-Renewal of Normal and Malignant Human Mammary Stem Cells. Cancer Res. 2006, 66, 6063–6071.
- Long, B.; Zhu, H.; Zhu, C.; Liu, T.; Meng, W. Activation of the Hedgehog Pathway in Chronic Myelogeneous Leukemia Patients. J. Exp. Clin. Cancer Res. 2011, 30, 8.
- Johnson, R.L.; Rothman, A.L.; Xie, J.; Goodrich, L.V.; Bare, J.W.; Bonifas, J.M.; Quinn, A.G.; Myers, R.M.; Cox, D.R.; Epstein, E.H.; et al. Human Homolog of Patched, a Candidate Gene for the Basal Cell Nevus Syndrome. Science 1996, 272, 1668–1671.
- Muzio, L.L. Nevoid Basal Cell Carcinoma Syndrome (Gorlin Syndrome). Orphanet J. Rare Dis. 2008, 3, 32.
- Dahmane, N.; Lee, J.; Robins, P.; Heller, P.; Ruiz I Altaba, A. Activation of the Transcription Factor Gli1 and the Sonic Hedgehog Signalling Pathway in Skin Tumours. Nature 1997, 389, 876–881.
- Goodrich, L.V.; Milenković, L.; Higgins, K.M.; Scott, M.P. Altered Neural Cell Fates and Medulloblastoma in Mouse Patched Mutants. Science 1997, 277, 1109–1113.
- Vořechovský, I.; Tingby, O.; Hartman, M.; Strömberg, B.; Nister, M.; Collins, V.P.; Toftgård, R. Somatic Mutations in the Human Homologue of Drosophila Patched in Primitive Neuroectodermal Tumours. Oncogene 1997, 15, 361–366.
- Tostar, U.; Malm, C.J.; Meis-Kindblom, J.M.; Kindblom, L.G.; Toftgård, R.; Undén, A.B. Deregulation of the Hedgehog Signalling Pathway: A Possible Role for the PTCH and SUFU Genes in Human Rhabdomyoma and Rhabdomyosarcoma Development. J. Pathol. 2006, 208, 17–25.
- Fan, L.; Pepicelli, C.V.; Dibble, C.C.; Catbagan, W.; Zarycki, J.L.; Laciak, R.; Gipp, J.; Shaw, A.; Lamm, M.L.G.; Munoz, A.; et al. Hedgehog Signaling Promotes Prostate Xenograft Tumor Growth. Endocrinology 2004, 145, 3961–3970.
- Tian, H.; Callahan, C.A.; Dupree, K.J.; Darbonne, W.C.; Ahn, C.P.; Scales, S.J.; De Sauvage, F.J. Hedgehog Signaling Is Restricted to the Stromal Compartment during Pancreatic Carcinogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 4254–4259.
- Ma, X.; Sheng, T.; Zhang, Y.; Zhang, X.; He, J.; Huang, S.; Chen, K.; Sultz, J.; Adegboyega, P.A.; Zhang, H.; et al. Hedgehog Signaling Is Activated in Subsets of Esophageal Cancers. Int. J. Cancer 2006, 118, 139–148.
- Zhao, C.; Chen, A.; Jamieson, C.H.; Fereshteh, M.; Abrahamsson, A.; Blum, J.; Kwon, H.Y.; Kim, J.; Chute, J.P.; Rizzieri, D.; et al. Hedgehog Signalling Is Essential for Maintenance of Cancer Stem Cells in Myeloid Leukaemia. Nature 2009, 458, 776–779.
- Turner, K.A. Assessment of a Potential Therapeutic Target in the Hedgehog Pathway for the Eradication of Primitive Chronic Myeloid Leukemia Cells. Ph.D. Thesis, University of British Columbia, Vancouver, BC, Canada, 2017.
- Wang, X.; Zhang, N.; Huo, Q.; Sun, M.; Dong, L.; Zhang, Y.; Xu, G.; Yang, Q. Huaier Aqueous Extract Inhibits Stem-like Characteristics of MCF7 Breast Cancer Cells via Inactivation of Hedgehog Pathway. Tumor Biol. 2014, 35, 10805–10813.
- Varnat, F.; Duquet, A.; Malerba, M.; Zbinden, M.; Mas, C.; Gervaz, P.; Ruiz I Altaba, A. Human Colon Cancer Epithelial Cells Harbour Active HEDGEHOG-GLI Signalling That Is Essential for Tumour Growth, Recurrence, Metastasis and Stem Cell Survival and Expansion. EMBO Mol. Med. 2009, 1, 338–351.
- Fu, P.; Zheng, X.; Fan, X.; Lin, A. Role of Cytoplasmic LncRNAs in Regulating Cancer Signaling Pathways. J. Zhejiang Univ. Sci. B 2019, 20, 1.
- Zhou, H.; Xiong, Y.; Peng, L.; Wang, R.; Zhang, H.; Fu, Z. LncRNA-CCSC1 Modulates Cancer Stem Cell Properties in Colorectal Cancer via Activation of the Hedgehog Signaling Pathway. J. Cell. Biochem. 2020, 121, 2510–2524.
- Wu, J.; Zhu, P.; Lu, T.; Du, Y.; Wang, Y.; He, L.; Ye, B.; Liu, B.; Yang, L.; Wang, J.; et al. The Long Non-Coding RNA LncHDAC2 Drives the Self-Renewal of Liver Cancer Stem Cells via Activation of Hedgehog Signaling. J. Hepatol. 2019, 70, 918–929.
- Guo, L.; Zhou, Y.; Chen, Y.; Sun, H.; Wang, Y.; Qu, Y. LncRNA ASAP1-IT1 Positively Modulates the Development of Cholangiocarcinoma via Hedgehog Signaling Pathway. Biomed. Pharmacother. 2018, 103, 167–173.
- Peng, W.; Wu, J.; Fan, H.; Lu, J.; Feng, J. LncRNA EGOT Promotes Tumorigenesis Via Hedgehog Pathway in Gastric Cancer. Pathol. Oncol. Res. 2017, 25, 883–887.
- Li, L.; Ma, T.T.; Ma, Y.H.; Jiang, Y.F. LncRNA HCG18 Contributes to Nasopharyngeal Carcinoma Development by Modulating MiR-140/CCND1 and Hedgehog Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10387–10399.
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66.




