
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Josu López-Fernández | + 11028 word(s) | 11028 | 2020-11-18 05:04:55 |
Video Upload Options
Lipases are biocatalysts with a significant potential to enable a shift from current pollutant manufacturing processes to environmentally sustainable approaches. The main reason of this prospect is their catalytic versatility as they carry out several industrially relevant reactions as hydrolysis of fats in water/lipid interface and synthesis reactions in solvent-free or non-aqueous media such as transesterification, interesterification and esterification. Because of the outstanding traits of Rhizopus oryzae lipase (ROL), 1,3-specificity, high enantioselectivity and stability in organic media, its application in energy, food and pharmaceutical industrial sector has been widely studied. Significant advances have been made in the biochemical characterisation of ROL particularly in how its activity and stability are affected by the presence of its prosequence. In addition, native and heterologous production of ROL, the latter in cell factories like Escherichia coli, Saccharomyces cerevisiae and Komagataella phaffii (Pichia pastoris), have been thoroughly described. Therefore, in this review, we summarise the current knowledge about R. oryzae lipase (i) biochemical characteristics, (ii) production strategies and (iii) potential industrial applications.
1. Introduction
Rhizopus oryzae is broadly employed in industry because it can carry out the synthesis of a great variety of products like organic acids (lactic and fumaric acids), volatile compounds and enzymes (cellulases, proteases, tannases, xylanasas, pyruvate decarboxylases, lipases etc.,) [1][2][3][4]. Concretely, according to Web of Knowledge data, R. oryzae lipase (ROL) is one of the most studied enzymes of this fungi. There are three major commercial formulations of this lipase (Table 1) and more than 200 scientific works have been published in the last 5 years highlighting the relevance of this enzyme. Therefore, the aim of this entry is to provide a complete overview of ROL in terms of biochemical properties, enzyme native and heterologous production and its industrial applications.
Table 1. Major commercial suppliers of Rhizopus oryzae lipase and some lipase properties [5].
|
Supplier |
Name |
Application |
Lipase Properties |
|
Amano |
Lipase DF “Amano” 15 |
Oil and fats |
Optimum pH range 6–7; stable pH range 4–7, optimum temperature range 35–40 °C, relatively specific to fatty acids |
|
Sigma |
Lipase from R. oryzae (no. 62305) |
Oil and fats |
Optimum pH 8, optimum temperature 40 °C |
|
Sigma |
Lipase, immobilised on Immobead 150 from R. oryzae (no. 89445) |
Pharmaceutical and bioenergy |
Optimum pH 7.5, optimum temperature 40 °C |
2. Biochemical Properties
R. oryzae lipase (ROL) is a protein synthesised as a precursor form containing a presequence of 26 amino acids, followed by a prosequence of 97 attached to the N-terminal of a 269 amino acids mature sequence (Figure 1) [6]. All known lipases from Rhizopus genus follow the same identical structure even though some amino acidic substitutions can be detected when their primary sequences are compared, not only between different species but also between different isolated strains of the same species (Figure 2). For instance, Ben Salah et al. [7] addressed the presence of several substitutions in the sequences of Rhizopus lipases published by his group and Sayari et al. [8], Beer et al. [6], Derewenda et al. [9] and Khono et al. [10].
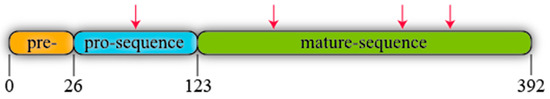
Figure 1. Schematic representation of R. oryzae lipase (ROL). Arrows stand for potential N-glycosylation points.
ROL contains four potential N-glycosylations sites (Figure 1) that follow the consensus sequence Asn-X-Ser/Thr, where X is any amino acid instead of proline. One of these putative sites is found in the prosequence where modifications in glycosylation patterns have been described to have an effect on protein secretion [11]. For instance, Yu et al. [12] added two extra N-glycosylation sites to ROL prosequence and expressed this mutant in Komagataella phaffii (Pichia pastoris). The extracellular activity and total protein were 218- and 6.25-fold higher respectively in the strain harbouring the two extra N-glycosylation sites than in the non-modified one highlighting the relevance of glycosylation.

Figure 2. Multiple alignment of the sequences published by (1) Beer et al. (ROL) [6], (2) Ben Salah et al. (ROL) [7], (3) Sayari et al. (ROL) [8], (4) Khono et al. (Rhizopus niveus lipase) [10] and (5) Derewenda et al. (Rhizopus delemar lipase) [9]. Matching amino acids are highlighted in yellow, mismatching in white. BLAST from U.S. National Library of Medicine and Snapgene have been used for the creation of this figure.
The presequence of ROL has been described to act as signal peptide promoting enzyme secretion, while the prosequence has been reported to exhibit diverse functions that are still under research. Beer et al. [13] depicted the significance of the latter in lowering lipase toxicity during its synthesis and in acting as intramolecular chaperone enabling the proper folding of the enzyme. In fact, genetically modified E. coli strains producing heterologous ROL without the prosequence resulted in cell lysis. To date, a large number of prosequences of different enzymes have also been identified to function as intramolecular chaperone and to assist the folding of their respective proteins [14]. In addition, several scientific works have related ROL prosequence with the translocation of the protein across the endoplasmic reticulum membrane, enhancement of free lipase stability and changes in enzyme substrate specificity. Nevertheless, the mechanisms that allow these traits are yet unknown despite the broad research carried out [15][16][17][18][19][20]. In any case, both the presequence and the prosequence are expected to be proteolytically removed to form the mature lipase. In spite of this, the native microorganism secretes a lipase that is attached to the N-terminal of mature sequence the 28 C-terminal amino acids of the prosequence (proROL), which then are cleaved via limited proteolysis catalysed by extracellular proteases [6][10][21]. However, some studies have indicated that the presence of these 28 amino acids of the prosequence alongside the mature sequence is enough for some of the presumed features of the prosequence to occur. For instance, higher free lipase stability and changes in enzyme specificity have been described when the 28 amino acids of the prosequence were expressed together with the mature sequence in K. phaffii [22]. In addition, these amino acids have also enabled lowering the toxicity of ROL production in E.coli [13] and they have been related to direct proteins to secretory pathway in Aspergillus oryzae [23].
The mature sequence of R. oryzae lipase (rROL) is constituted by 269 amino acids and the protein formed by them has a molecular weight (MW) of 29.542 kDa and a isoelectric point (pI) of 8—calculated by Expasy Proteomics Server [7]. These results agree with the published experimental data (Table 2) in which MW and pI values around 29 kDa and 8 have been respectively reported [8][10][24][25]. However, variations in these values can be found because of the presence of the 28 amino acids of the prosequence described above [22][10][20][26][27]. In this case, MW increases to 32 kDa and pI decreases roughly to 7, highlighting the average acid nature of these 28 amino acids. Besides, the production of a lipase including the whole prosequence and close to 40 kDa has also been described (entire-proROL) [6][15].
Table 2. Biochemical properties and substrate specificity of different published works dealing with ROL.
|
Lipase Name 1 |
MW (kDa) |
Isoelectric Point |
pH Optimum |
T Optimum (°C) |
Substrate Specificity |
Ref. |
|
rROL |
29 |
|
8/7.252 |
30/40 2 |
C12>C10>C8>C4 4 |
[22] |
|
proROL |
32 |
|
7.25 |
40 |
C8>C12>C10>C4 4 |
[22] |
|
rROL |
30 |
|
8.5 |
|
|
[6] |
|
entire-proROL |
40 |
|
8 |
|
|
[6] |
|
pre-entire-proROL 3 |
42 |
|
8 |
|
|
[6] |
|
rROL |
29 |
|
8 |
37 |
|
[7] |
|
rROL |
29 |
|
|
|
|
[8] |
|
proROL |
32 |
|
|
|
|
[8] |
|
proROL |
34 |
|
6–6.5 |
35 |
|
[10] |
|
rROL |
30 |
|
6 |
40 |
|
[10] |
|
proROL |
35 |
|
9 |
40 |
C16>C18>C12>C8>C4 5 C16>C12>C8>C18>C4 6 |
[18] |
|
proROL |
32 |
6.9 |
|
|
|
[20] |
|
rROL |
30 |
9.3 |
8.25 |
30 |
C8>C10>C6>C4>C12>C16,C14>C2 6 |
[24] |
|
proROL |
35 |
|
5.2 |
30 |
C12>C10>C8>C6>C16>C5>C4>C3>C2 4 |
[25] |
|
proROL |
32 |
7.6 |
7.5 |
35 |
C8>C6>C4>C2 6 |
[26] |
|
rROL |
29 |
|
|
|
C12>C10>C8>C6>C4>C3>C2 4 C8>C10>C18>C4>C6 6 |
[28] |
|
proROL |
34 |
|
|
|
C2>C3>C8>C6>C12>C10>C4 4 C8>C10>C4>C6>C18 6 |
[28] |
|
proROL |
|
|
8 |
40 |
|
[29] |
|
rROL |
30.3 |
8.6 |
8–8.5 |
30 |
|
[30] |
|
proROL |
|
|
8.5 |
30 |
|
[31] |
|
proROL |
37 |
|
8.5 |
40 |
|
[32] |
|
rROL |
29 |
|
8 |
|
|
[33] |
|
ROL |
17 |
4.2 |
7 |
40 |
|
[34] |
|
ROL |
|
|
7 |
40 |
|
[35] |
|
ROL |
|
|
6 |
45 |
C8>C4>C6>C2 6 C8>C12>C14>C16>C18 5 |
[36] |
|
proROL |
32 |
|
7 |
35 |
|
[37] |
|
ROL |
|
|
6 |
30 |
C7,C8,C12,C16>C2,C3,C4,C18 5 |
[38] |
|
ROL |
|
|
7.5 |
50 |
|
[39] |
|
proROL |
32 |
|
7.5 |
30–40 |
|
[40] |
|
ROL |
14.45 |
6.5 |
9 |
30–40 |
C16>C18>C12>C8>C4>C2 4 |
[41] |
|
|
|
|
8.3 |
35–37 |
|
[42] |
|
proROL |
35 |
|
|
|
C10>C14>C12>C8>C6>C4>C16 6 |
[15] |
|
entire-proROL |
46 |
|
|
|
|
[15] |
1Names are based on the established nomenclature in this review. ROL indicates that the lipase cannot be classified under the determined parameters in this work; 2 different values caused by the employment of 200 or 400 mM tris-HCl buffer; 3 pre-entire-proROL includes the presequence as well as entire-proROL; 4 p-nitrophenol esters were employed for substrate specificity analysis; 5 methyl esters of different carbon chain length were employed for substrate specificity analysis. Saturated methyl esters are just considered; 6 homotriacylglycerols were employed for substrate specificity analysis.
The 3D structure of the lipase from R. oryzae [9][10] (Figure 3) and several microorganisms more, such as Geotrichum candidum [43], Candida rugosa [44], Pseudomonas glumae [45] and Penicillium camemberti [46] have been crystallographically resolved and showed that all lipases have a common α/β hydrolase fold structure that can also be found in other hydrolases. Regarding ROL, it contains nine α-helixes and eight β-strands forming a molecule that it is stabilised by three disulfide bonds between residues 29–269, 40–43 and 235–244 [10]. In addition, this structure contains three key components that can be also found in most lipases besides ROL, the lid, the active site and the oxyanion hole [47]. The lid is an amphiphilic loop—also called flap—that covers the active site preventing the access of the substrate while the enzyme is in aqueous medium [48]. The active site, in turn, is mainly responsible for carrying out enzyme catalysis and consists, in all α/β hydrolases, of a highly conserved catalytic triad formed by a nucleophilic, a catalytic acidic and a histidine residues. In lipases, this triad is composed of nucleophilic serine residue and an aspartic or glutamic acid residue that it is bonded to a histidine; hence, lipases are classified as serine hydrolases. In the specific case of ROL, the lid domain is a short α-helix structure formed by six amino acids (FRSAIT) and the active site is formed by three amino acids Ser145, Asp204 and His257 [9][10][49][50]. The function of these two elements is crucial during catalysis in which the lipase binds to the water/lipid interface and the lid opening occurs by a concomitant structural change in the substrate-binding site that enables the coupling of the substrate to the active site—lid-closed and partially opened 3D structures of Rhizopus delemar (=oryzae) lipase have been described by Derewenda et al. [9]. The structural change undergone is known as ‘interfacial activation’ and it is a unique property of lipases that enables them to hydrolyse insoluble esters and to distinguish them from esterases that can hydrolyse water-soluble esters [47][51][52][53]. It must be highlighted that the 28 amino acids of the prosequence introduced above have been deemed to interfere in this process as they are located next to the lid region and contain 50% hydrophobic residues. Therefore, this sequence extends the hydrophobic patch created in the open lipase by the open lid and the catalytic crevice influencing the interaction with the lipidic substrate [8]. This role might explain some of the assumed properties of these 28 amino acids, however, the mechanism remains unknown. Additionally, together with the catalytic triad and the lid, the oxyanion hole plays an important role and it is also a highly conserved sequence that largely influences the catalytic efficiency of the enzyme. During the hydrolysis reaction, a negatively charged tetrahedral intermediate is generated and it gets stabilised by hydrogen binding with the oxyanion hole [47][54][55]. This function has been described to be presumably performed by the hydroxyl and main-chain amide groups of Thr83 in ROL [9][10][56].

Figure 3. Three-dimensional structure of R. delemar (= oryzae) lipase from two different points of view. PDB ID: 1TIC. Image obtained from iCn3D web-based 3D structure viewer.
Due to the relevance of the lipolytic activity of this enzyme, it has been widely researched in order to know how it is affected by the conditions of reaction medium. Guillen et al. [28] described that ionic strength has a remarkable impact. Actually, the relative activity of ROL in 200 mM Tris-HCl was reported to be twice the activity observed in 400 mM. Moreover, as all enzymes, ROL activity is highly influenced by the pH and temperature. Optimum activity pH values of 8 have been principally reported [6][7][24][26][28][57][29][30][31][32][33]. However, other studies have also stated more acid [25][58][35][36][37][38][39][59] and basic [18][41][42] optimums. Regarding temperature, most of the published optimum values can be found between 30 and 45 °C. In fact, 40 °C has been the most commonly reported optimum [22][10][18][28][29][32][58][35][59][41] although lower [7][10][25][26][28][30][31][37][38] and higher [36][39] values have also been described. Nevertheless, for both pH and temperature, as can be observed in Table 2, some of the differences are based on the presence of the 28 amino acids of the prosequence. In this line, Kohno et al. [10] reported these differences and afterwards, other works [22][60][28] described similar results highlighting the relevance of these amino acids in lipase catalytic performance.
The presence of metal ions in reaction medium has been extensively studied as they play different and important roles influencing the structure and activity of enzymes. These ions may bind to some of the amino acid side chains of the lipase and participate in catalysis, interfere with the bonds between amino acid side chains and cause denaturation of the active site or alter enzyme activity by stabilising or destabilising enzyme conformation [37][61][62][63]. Amongst the different published works some contradictory information can be found. Nevertheless, there are some metal ions that have been clearly described to enhance or worsen ROL and other lipases performance. Wang et al. [18] and other authors [34][37][41] found that Ca2+ increases ROL activity as it might create electrostatic interactions that mask the repulsions either between the enzyme and its emulsified substrate or between the enzyme and product-free fatty acids [30]. On the other hand, Hg2+ has been reported to act like a ROL activity inhibitor suggesting that thiol groups are required for the adequate function of the enzyme [25]. Similar results have been reported with other lipases from Pseudomonas aeruginosa AAU2 [64], Galactomyces geotrichum Y05 [65], Yarrowia lipolytica [66] and Candida rugosa [67]. In addition, no significant effects have been observed with the chelating agent EDTA, indicating that ROL activity is independent of any metal, hence, it is not a metalloprotein [25][41].
ROL activity has also been analysed in presence of amino-acid-modifying agents in order to elucidate the relevance of the different amino acids in protein catalytic performance. N-Bromosuccinimide (NBS), which acts over tryptophan residues, has been reported to strongly inhibit enzyme activity indicating that the protein might have a tryptophan residue involved in its activity [25][34]. In the case of phenylmethylsulfonyl fluoride (PMSF), a serine protease inhibitor whose activity is related to serine residues modification, no clear results have been reported. Kantak et al. [34] indicated that this agent has a relevant effect while Hiol et al. [26] stated exactly the opposite. However, these differences might be caused by the different disposition of the lipase lid during the assay, that is, if it was open or not, it could allow or not the interaction of PMSF with the serine residue of the active site [68].
ROL activity—as most lipases from Rhizopus genus—has a strong 1,3-regiospecificity that makes its activity interesting for several industrial processes such as fat and oil modification for structured lipids production [5][4][26][36]. Nevertheless, Li et al. [69] reported, while studying ROL methanolysis performance, that this lipase was not regiospecific although showed a preference to 1,3-positions. These results were lately confirmed with Rhizopus arrhizus (=oryzae) lipase [70]. However, Okumura et al. and Song et al. [38][71] stated that Rhizopus delemar (=oryzae) and R. oryzae lipases, respectively, do not hydrolyse the ester bond in position 2. Afterwards, Canet et al. and Cao et al. [72][73] proved that mature ROL exhibits a negligible activity on 2-monoolein highlighting that the lipase has a strong 1,3-regioespecificity. The observed dissimilarities amongst different works might be due to the different employed reaction conditions that could enhance spontaneous acyl-migration, or the presence of the 28 amino acids of the prosequence that has already been described to have an effect on lipase specificity [22][13]. Besides 1,3-regiospecificity, substrate specificity of ROL has been also widely studied. Many of the published works are based on the employment of p-nitrophenol esters of different carbon-chain length. For instance, ROL isolated and characterised by Adak et al. [41] was reported to be more specific to long carbon-chain p-nitrophenol esters, concretely to p-nitrophenol palmitate (C16). Guillen et al. [28] reported a similar trend for rROL produced in K. phaffii and, although a higher specificity to short carbon-chain p-nitrophenol esters was detected for proROL, the presence of esterases in the commercial product was concluded to be the reason. In fact, Tako et al. [25] also observed that the longer the carbon-chain, the higher the specificity of ROL. However, in this last case, the maximum was obtained with p-nitrophenol dodecanoate (C12) and not palmitate. ROL substrate specificity has been also analysed with homotriacylglycerols, that is, triacylglycerols in which the three fatty acids are identical. C8 and C10 homotriacylglycerols—triacylglycerols containing three C8 and C10 fatty acids respectively—are preferably hydrolysed by ROL while it barely acts over C2 and C4 homotriacylglycerols. In contrast to some of the published works, some authors have also described that no significant differences were observed with those substrates between rROL and proROL [15][24][26][28].
Lipases are widely known for their capacity to carry out synthesis reactions in non-aqueous mediums. In fact, as previously mentioned, this capacity makes them relevant for many industrial processes in which these reactions are needed, or the solubility of substrates/products requires the use of organic solvents. Therefore, the higher the lipase stability in these solvents, the more suitable the lipase for industrial applications [74]. ROL has been extensively described as a tolerant enzyme to non-aqueous solvents [18][26][31], particularly in alkanes and long-chain alcohols such as hexane and dodecanol respectively. However, polar solvents like acetone or short-chain alcohols have an important negative effect on the enzyme because they strip off the crucial bound water from the enzyme’s surface [75]. In some cases, it is remarkable the different results that can be obtained between the stability of the enzyme in a solvent, such as methanol and ethanol, and the operational stability employing that solvents as substrate. For instance, methanol has proven to be more detrimental than ethanol during biodiesel synthesis while during stability assays exactly the opposite result was obtained [22].
3. Rhizopus oryzae Lipase Production and Bioprocess Engineering
First attempts of ROL production were made with the original fungi isolated from palm fruit [26][31]. R. oryzae secretes, as previously mentioned, one form of lipase with a molecular weight close to 32 kDa—the mature sequence including 28 amino acids of its prosequence. However, a second form of ROL with a molecular weight around 29 kDa was detected after keeping the supernatant at 6 °C for few days; i.e., the lipase form corresponding to the loss of the 28 amino acids [8]. Consequently, the distinct lipases derived from R. oryzae described in the literature are originated because of the different proteolytic processing and not because of the presence of different genes [6].
To increase ROL industrial production, its expression in a cell factory is mandatory. This way, production cost, bioprocess engineering and downstream complexity are minimised [5].
In Escherichia coli, the presence of disulphide bonds in ROL structure and the lack of the necessary enzymes to process fungal maturation signals were the main causes that led to the production of enzymatically inactive protein as insoluble aggregates [6]. Thereafter, active lipase was obtained at lab scale by subjecting these aggregates to a refolding process. However, the large-scale production was not implemented due to the high cost of the procedure [13]. Despite that, Di Lorenzo et al. [76], achieved the production of an active and soluble ROL and proROL using the E. coli Origami (DE3) strain and pET-11d expression system. The final specific activities of both enzymes were quite similar but the yield of proROL production was higher than ROL, likely because of the toxic effect of the latter towards the host cells.
To avoid the inherent problems of prokaryotic cell factories producing eukaryotic proteins, particularly those related to post-translational processing, eukaryotic cell factories were tested for ROL production.
The extracellular production of ROL has been studied in S. cerevisiae and K. phaffii (P. pastoris) by expressing essentially three different genes. A gene encoding the prosequence of 97 amino acids fused to the N-terminal of the mature lipase region of 269 amino acids (proROL-gene), a gene encoding a truncated prosequence of its 28 C-terminal amino acids fused to the N-terminal of the mature lipase region (28proROL-gene) and a gene encoding the mature lipase (rROL-gene). Regardless of proROL-gene or 28proROL-gene expression, a protein with only 28 amino acids of the prosequence plus the mature lipase (proROL) was detected. Exceptionally, the complete prosequence plus the mature lipase region (entire-proROL) was also reported with proROL-gene construction. With respect to the rROL-gene construction, jus the mature lipase (rROL) was obtained.
First attempts of producing ROL in eukaryotic platforms were made with the widely used cell factory S. cerevisiae. Takahasi et al. [15] reported that S. cerevisiae secreted two active lipases when it was transformed with the proROL-gene fused to the pre-α-factor, the entire-proROL and proROL—the lipase formed after Kex2-like protease cleavage of the prosequence. In parallel, when S. cerevisiae strains were transformed with rROL-gene fused to the pre-α or prepro-α factor encoding gene, almost no activity was detected, highlighting the mentioned relevance of ROL prosequence during lipase production [16][19][21].
A summary of the results obtained with these cell factories is shown in Table 3.
Table 3. Summary of E. coli and S. cerevisiae cell factories expressing Rhizopus oryzae lipase.
|
Cell Factory |
Promotor/Vector |
Lipase |
Production |
Lipolytic Activity |
Reference |
|
E. coli Origami DE3 |
pET11 |
proROL |
Intracellular |
166 U mL−1 |
[76] |
|
pET22 |
proROL |
Intracellular |
82 U mL−1 |
||
|
S. cerevisiae |
UPR-ICL |
rROL |
Extracellular |
0.29 U flask−1 |
[15] |
|
UPR-ICL |
proROL |
Extracellular |
191 U flask−1 |
3.1. Komogataella phaffii Cell Factory
Unlike the reported results with S. cerevisiae, when proROL-gene was expressed in K. phaffii cell factory, only proROL was detected in the medium, which might indicate that the activity of the Kex2-like protease is higher in this cell factory than in S. cerevisiae [17]. Moreover, rROL-gene was satisfactorily expressed and the corresponding lipase was detected in the supernatant [24].
This appropriate performance on ROL secretion, jointly with the well-known excellent characteristics of K. phaffii, make this yeast the most suitable cell factory for heterologous ROL production [77][78][79][80]. In addition, K. phaffi does not produce endogenous extracellular lipases or esterases [81]. Thus, downstream processes might be easier and cheaper. However, two of the bottlenecks of K. phaffii cell factory are transformed clones screening and selecting the best operational strategy to maximise production. To minimise this problem, the use of microbioreactor devices has been successfully implemented [82]. Further information about K. phaffii as cell factory for ROL production was summarised by López-Fernández et al. [83]
3.2. Whole cells
Hama et al. reported that rROL and proROL are located in different regions in R. oryzae cells, proROL in the cell wall and rROL bound to the cell membrane. Besides, these cells have been successfully employed as whole cells biocatalysts (WCB) in many relevant biotransformations, for instance, for enzymatic biodiesel production [84][85]. It must be highlighted that the fatty acid composition of the membrane has been reported to influence lipase activity and stability during biodiesel reactions [86].
Modified S. cerevisiae strains producing ROL have also been used as WCB [87][88]. Matsumoto et al. [87] reported the intracellular production of proROL in S. cerevisiae under the 5′ upstream region of the isocitrate lyase gene of Candida tropicalis (UPR-ICL). Additionally, the expression of the lipase under the constitutive promoter glyceraldehyde-3-phosphate dehydrogenase was also studied. However, this system did not improve the results obtained with UPR-ICL.
proROL was successfully expressed under PAOX control and displayed on Mut+ phenotype K. phaffii cell surface using the Flo1P anchor system previously developed in S. cerevisiae. The obtained WCB showed higher thermal stability than free enzyme [89]. Additionally, a similar approach using Sed1p anchor protein was studied in a MutS phenotype. In the same sense, the obtained biocatalysts was stable in a wide range of temperatures and pH [90].
4. Industrial Applications of Rhizopus oryzae Lipase
Its 1,3-regiospecificity and catalytic versatility make ROL appropriate for improving the sustainability of food, pharmaceutical and energy industry [5][91][92].
4.1. Biodiesel Production
Because of petroleum depletion and environmental concerns, in the past decade, biodiesel (mono-alkyl esters of long chain fatty acids) is gathering significant interest as a renewable, biodegradable and more environmentally friendly alternative to fossil fuels. Biodiesel can be classified into three different generations based on the source from which it is derived. First-generation biodiesel is synthesised with edible-oils such as soybean or sunflower oils. Therefore, it might cause the so-called “food vs. fuel” ethical issue because of the use of food and agricultural lands for biofuel production [93]. In order to prevent this problem, alternative substrates have emerged for biodiesel synthesis, leading to second- and third-generation biodiesel production. The former uses non-edible oils that are not considered for human consumption and are produced from crops that, even if they require lands, are generally poor lands not useful for agriculture. Meanwhile, third-generation biodiesel completely avoids ethical issues by using microbial lipids and oleaginous wastes such as oils from microalgae or oleaginous yeasts and waste cooking oils (WCO) respectively [94][95][96][97]. Additionally, there is a fourth-generation biodiesel that is at its preliminary research stages and is based on man-made biological tools, that is, biodiesel producing genetically modified microorganisms [98][99].
Typically, these alternative substrates, those yielding second- and third-generation biodiesel, have a higher free fatty acid (FFA) content, which can make biodiesel production through chemical synthesis—the most common process for current industrial biodiesel production—more complex because a previous operation of FFA neutralisation is required to avoid soap formation, an usual side reaction when substrates with high FFA content and basic catalysts are employed [95][100][101]. In this context, enzymatic biodiesel synthesis with lipases arouses as an alternative owing to its several advantages such as the milder reaction conditions, less water consumption, easier downstream and particularly, the absence of side reactions and consequently the capacity of employing substrates with high FFA content [102][103]. In fact, substrates with initial high amounts of FFA have been reported to enhance enzymatic biodiesel synthesis reaction rate and biocatalysts operational stability [102][104][105]. Given all the advantages, numerous lipases have been studied in this biotransformation with significant results, such as the lipases of Candida rugosa [106][107], Candida antartica [108][109] and Burkholderia cepacia [110][111].
In the seeking for the best lipase to make enzymatic biodiesel feasible at industrial scale, lipases’ regiospecificity has become a crucial trait. Non-specific enzymes produce mono-alkyl esters and glycerol, which is an undesired by-product of the transesterification reaction that has been described to hinder reaction progress or even affect negatively on enzymes stability and biodiesel downstream [112]. Conversely, 1,3-regioespecific lipases, avoid glycerol formation by producing 2-monoacylglycerol which acts as lubricant and in certain amount, upgrades biodiesel characteristics [113][114][115]. Furthermore, monoacylglycerols can improve the cost-effectiveness of a biodiesel biorefinery as they are more valuable products than glycerol because of their utility in pharmaceutical and food industry as emulsifiers [116][117][118]. Consequently, ROL has been widely studied in biodiesel production because of its regiospecificity.
Considering biodiesel ethical issues, even if several studies have employed ROL with edible oils such as olive [119], rapeseed [120][121], soybean [122][123][124][125] and sunflower [126][127] oils—commonly as model substrates for research—most of the published works have focused on the use of alternative substrates (Table 4). Jatropha curcas oil is one of the non-edible oils with higher potential for second-generation biodiesel production, probably because of the easy cultivation process and worldwide spread of the plant [128]. Rodrigues et al. [129] reported yields close to the theoretical 100%—real 66% considering ROL 1,3-regioespecificity—and high operational stability of the biocatalysts. In Table 4 are detailed other studies with promising results using this substrate as well as other non-edible oils like Pistacia chinensis bge oil [130], Tung oil [32], Calophyllum inophyllum oil [131] and alperujo oil (olive pomace) [115].
Table 4. Summary of biodiesel production with Rhizopus oryzae lipase as main biocatalyst.
|
Substrates |
Lipase |
Immobilisation Technique |
Reactor Type |
Stepwise Addition |
Biodiesel Generation |
Yield-Conversion/Op. Stability |
Ref. |
|
|
OO + MeOH |
rROL |
IA onto ReliZymeTM OD 403M |
PBR |
Yes |
1st |
Y: PBR 49.1% OS: second batch 44.8% |
[119] |
|
|
OO + MeOH |
rROL |
IA onto ReliZymeTM OD 403M |
STR |
Yes |
1st |
Y: STR 33.56% OS: second batch 7.7% |
[119] |
|
|
RO + MeOH |
proROL |
WCB over agar plate |
SLLB |
No |
1st |
No biodiesel production |
[120] |
|
|
RO + EtOH |
proROL |
WCB over agar plate |
SLLB |
No |
1st |
No biodiesel production |
[120] |
|
|
RO + MeOH |
proROL |
WCB over agar plate |
SGLB |
No |
1st |
Y: 58% |
[120] |
|
|
RO + EtOH |
proROL |
WCB over agar plate |
SGLB |
No |
1st |
Y: 72% |
[120] |
|
|
Crude CO + MeOH |
proROL |
Free enzymes |
BR |
Yes |
1st |
Y: 68.56% |
[121] |
|
|
Crude CO + MeOH |
proROL-CRL |
Free enzymes |
BR |
Yes |
1st |
Y: 84.25% |
[121] |
|
|
Crude CO + MeOH |
proROL-CRL |
CI onto functionalised silica gel |
BR |
Yes |
1st |
Y: 88.9% |
[121] |
|
|
SYO + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
1st |
Y: 82.2% OS: after 6 cycles almost all activity loss |
[122] |
|
|
SYO + MeOH |
proROL |
CI WCB immobilised onto BSPs |
BR |
Yes |
1st |
Y: 92.2% OS: after 6 cycles no loss of activity |
[122] |
|
|
SYO + EtOH |
proROL |
IA onto microporous resin NKA (polystyrene) |
BR |
Yes |
1st |
Y: 58.5% |
[123] |
|
|
SYO + EtOH |
proROL-CRL |
IA onto microporous resin NKA (polystyrene) |
BR |
Yes |
1st |
Y: 80.8% |
[123] |
|
|
SYO + EtOH |
proROL-Novozyme 435 |
proROL: IA onto microporous resin NKA (polystyrene). Novozyme 435: IA onto Lewatit VP OC 1600 |
BR |
Yes |
1st |
Y: 98.5% OS: after 20 cycles Y decreased to 78.3% |
[123] |
|
|
SYO + EtOH |
proROL-PFL |
IA onto microporous resin NKA (polystyrene) |
BR |
Yes |
1st |
Y: 55.8% |
[123] |
|
|
SYO + MeOH |
proROL |
CI onto magnetic chitosan microspheres |
MSFBR |
Yes |
1st |
Y: 91.3% OS: after 6 reaction cycles Y decreased to around 80% |
[124] |
|
|
SYO + MeoH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
1st |
Y: over 90% OS: after 10 reaction cycles Y decreased to 10% |
[125] |
|
|
SYO + MeoH |
proROL |
WCB immobilised into BSPs |
PBR |
Yes |
1st |
Y: over 90% OS: after 10 reaction cycles Y decreased to 80% |
[125] |
|
|
SO + EtOH |
proROL |
CI onto modified sepiolite with p-hydroxybenzaldehyde linker |
BR |
No |
1st |
C: 84.3% OS: after 9 cycles C decreased to 21.4% |
[126]] |
|
|
SO + EtOH |
proROL |
CI onto modified sepiolite with benzylamine-terephthalic aldehyde linker |
BR |
No |
1st |
|
[126] |
|
|
SO + EtOH |
proROL |
IE onto demineralised sepiolite |
BR |
No |
1st |
Y: 90.2% OS: proROL IE after 9 cycles C decreased to 18.1% |
[126] |
|
|
Pistacia chinensis bge seed oil + MeOH |
rROL |
CI onto Amberlite IRA-93 |
BR |
Yes |
2nd |
Y: 92% OS: after 8 cycles Y decreased to 60% |
[130] |
|
|
Pistacia chinensis bge seed oil + MeOH |
rROL |
IA microporous resin HPD-400 |
BR |
Yes |
2nd |
Y: 94% OS: after 8 cycles Y decreased to 50% |
[130] |
|
|
Calophyllum inophyllum linn oil + MeOH |
proROL |
WCB immobilised into BSPs |
PBR |
Yes |
2nd |
Y: 92% OS: after 6 cycles Y decreased a 4.9% |
[131] |
|
|
Oil extracted from Nannochloropsis gaditana + MeOH |
proROL |
WCB |
BR |
Yes |
3rd |
Y: 83% OS: after 3 cycles Y decreased to 71% |
[132] |
|
|
Oil extracted from Nannochloropsis gaditana + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
3rd |
Y: 70% OS: second cycle Y decreased to 43% |
[132] |
|
|
Oil extracted from Nannochloropsis gaditana + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
3rd |
Y: 83% OS: after 3 cycles Y decreased to 71% |
[133] |
|
|
Oil extracted from Nannochloropsis gaditana + MeOH |
proROL |
WCB |
TPB |
No |
3rd |
Y: 58% |
[134] |
|
|
Oil extracted from Nannochloropsis gaditana + EtOH |
proROL |
WCB |
TPB |
No |
3rd |
Y: 92% |
[134] |
|
|
Oil extracted from Botryococcus braunii + MeOH |
proROL |
WCB |
TPB |
No |
3rd |
Y: 58% |
[134] |
|
|
Oil extracted from Botryococcus braunii + EtOH |
proROL |
WCB |
TPB |
No |
3rd |
Y: 68% |
[134] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
Free enzyme |
BR |
Yes |
3rd |
C: 75% |
[135] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
IA onto MNP |
BR |
Yes |
3rd |
C: 46% OS: after 5 cycles decreased to 10% |
[135] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
CI onto AP modified MNP |
BR |
Yes |
3rd |
C: 53% OS: after 5 cycles C decreased to 25% |
[135] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
CI onto AP-GA modified MNP |
BR |
Yes |
3rd |
C: 69.8% OS: after 5 cycles C decreased to 45% |
[135] |
|
|
Sludge palm oil + MeOH |
proROL |
IE into alginate-polyvinyl alcohol beads |
BR |
No |
3rd |
Y: 91.30% OS: no activity loss after 15 cycles |
[136] |
|
|
Oil extracted from SCG + MeOH |
R. delemar (= oryzae) lipase |
Free enzyme |
BR |
No |
3rd |
Y: 18% |
[137] |
|
|
WCO + MeOH |
proROL |
Free enzyme |
BR |
|
3rd |
Y: 93% |
[138] |
|
|
WCO + iso-propanol |
proROL |
Free enzyme |
BR |
|
3rd |
Y: 86.8% |
[138] |
|
|
WCO + iso-butanol |
proROL |
Free enzyme |
BR |
|
3rd |
Y: 80.2% |
[138] |
|
|
WCO + iso-amyl alcohol |
proROL |
Free enzyme |
BR |
|
3rd |
Y: 64% |
[138] |
|
|
WCO + MeOH |
proROL |
WCB IE into calcium alginate beads |
BR |
|
3rd |
Y: 84% |
[138] |
|
|
WCO + iso-propanol |
proROL |
WCB IE into calcium alginate beads |
BR |
|
3rd |
Y: 71% |
[138] |
|
|
WCO + iso-butanol |
proROL |
WCB IE into calcium alginate beads |
BR |
|
3rd |
Y: 62% |
[138] |
|
|
WCO+ iso-amyl alcohol |
proROL |
WCB IE into calcium alginate beads |
BR |
|
3rd |
Y: 43% |
[138] |
|
|
JO + MeOH |
proROL |
WCB IE into sodium alginate beads |
BR |
No |
2nd |
Y: 80.5% OS: after 6 cycles Y decreased to 61.5% |
[139] |
|
|
KO + MeOH |
proROL |
WCB IE into sodium alginate beads |
BR |
No |
2nd |
Y: 78.3% OS: after 6 cycles Y decreased to 63.4% |
[139] |
|
|
SYO + MeOH |
proROL |
WCB |
BR |
Yes |
1st |
Y: 80% OS: after 3 cycles Y decreased to 18% |
[140] |
|
|
SYO + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
1st |
Y: 82% OS: after 10 cycles Y decreased to 10% |
[140] |
|
|
SYO + MeOH |
proROL |
CI WCB immobilised into BSPs |
BR |
Yes |
1st |
Y: 74% OS: after 35 cycles Y decreased to 65% |
[140] |
|
|
SYO + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
1st |
Y: 82% OS: after 6 cycles Y decreased to 48% |
[141] |
|
|
SYO + MeOH |
proROL |
CI WCB immobilised into BSPs |
BR |
Yes |
1st |
Y: 80% OS: after 6 cycles Y decreased to 70% |
[141] |
|
|
ALO + MeOH |
rROL |
IA onto rice husk |
BR |
Yes |
2nd |
|
[142] |
|
|
ALO + MeOH |
rROL |
IA onto ReliZymeTM OD403 |
BR |
Yes |
2nd |
Y: 64.5% OS: after 7 cycles Y decreased to 41.3% |
[142] |
|
|
Crude microbial oil from Candida sp. LEB-M3 + MeOH |
rROL |
IA onto ReliZymeTM OD403 |
BR |
Yes |
3rd |
Y: 38% OS: after 7 cycles Y decreased to 26.6% |
[143] |
|
|
Neutralised microbial oil from Candida sp. LEB-M3 + MeOH |
rROL |
IA onto ReliZymeTM OD403 |
BR |
Yes |
3rd |
Y: 38% |
[143] |
|
|
OO + MeOH |
rROL |
IA onto ReliZymeTM OD403 |
BR |
Yes |
1st |
Y: 54.3% OS: after 7 cycles Y decreased to 40% |
[143] |
|
|
OA + MeOH |
rROL |
IA onto ReliZymeTM OD403 |
BR |
Yes |
1st |
Y: 68% |
[143] |
|
|
RO + EtOH |
proROL |
IA onto microporous resin NKA |
BR |
No |
1st |
Y: above 98% OS: After 10 cycles Y decreased to 60% |
[144] |
|
|
JO + MeOH |
proROL-CRL |
WCB (proROL) and free enzyme (CRL) IE into sodium alginate beads |
PBR |
No |
2nd |
Y: 84.2% |
[145] |
|
|
KO + MeOH |
proROL-CRL |
WCB (proROL) and free enzyme (CRL) IE into sodium alginate beads |
PBR |
No |
2nd |
Y: 81% |
[145] |
|
|
WCO + MeOH |
proROL |
WCB IE into sodium alginate beads |
BR |
No |
3rd |
Y: 94.01% |
[146] |
|
|
WCO + Methyl acetate |
proROL |
WCB IE into sodium alginate beads |
BR |
No |
3rd |
Y: 91.11% |
[146] |
|
|
WCO + Ethyl acetate |
proROL |
WCB IE into sodium alginate beads |
BR |
No |
3rd |
Y: 90.06 |
[146] |
|
|
WCO + MeOH |
proROL |
IE into sodium alginate beads |
BR |
No |
3rd |
Y: 83% |
[146] |
|
|
WCO + Methyl acetate |
proROL |
IE into sodium alginate beads |
BR |
No |
3rd |
Y: 80% |
[146] |
|
|
WCO + Ethyl acetate |
proROL |
IE into sodium alginate beads |
BR |
No |
3rd |
Y: 78% |
[146] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
IA into MNP |
BR |
Yes |
3rd |
Y: 45% OS: after 5 cycles Y decreased to 10% |
[147] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
IA into MGO |
BR |
Yes |
3rd |
Y: 51% OS: after 5 cycles Y decreased to 16% |
[147] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
IA into MGO-AP |
BR |
Yes |
3rd |
Y: 54% OS: after 5 cycles Y decreased to 25% |
[147] |
|
|
Oil extracted from Chlorella vulgaris + MeOH |
proROL |
CI into MGO-AP-GA |
BR |
Yes |
3rd |
Y: 68% OS: after 5 cycles Y decreased to 58.77% |
[147] |
|
|
Cottonseed oil + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
1st |
Y: 27.9% |
[148] |
|
|
Rubber seed oil + MeOHe |
proROL |
Free enzyme |
BR |
Yes |
2nd |
Y: 31% |
[149] |
|
|
Rubber seed oil + Ethyl acetate |
proROL |
Free enzyme |
BR |
No |
2nd |
Y: 33.3% |
[149] |
|
|
SYO + MeOH |
proROL-CRL |
CI onto silica gel pretreated with AP and GA |
BR |
Yes |
1st |
Y: 99.99% OS: after 20 cycles Y decreased to 85% |
[150] |
|
|
RO deodoriser distillate + MeOH |
proROL |
Free enzyme |
BR |
Yes |
1st |
Y: 93.07% |
[151] |
|
|
RO deodoriser distillate + MeOH |
proROL-CRL |
Free enzyme |
BR |
Yes |
1st |
Y: 98.16% |
[151] |
|
|
ALO + MeOH |
rROL |
CI onto ET, AP and GA pretreated ReliZymeTM HFA403 |
BR |
Yes |
2nd |
Y: 57.16% OS: after 5 cycles Y decreased a 12.31% |
[115] |
|
|
ALO + EtOH |
rROL |
CI onto ET, AP and GA pretreated ReliZymeTM HFA403 |
BR |
Yes |
2nd |
Y: 60.25% OS: after 7 cycles Y decreased a 11.89% |
[115] |
|
|
Triolein + MeOH |
rROL |
Free enzyme |
BR |
No |
1st |
Y: 71.2% |
[72] |
|
|
Triolein + EtOH |
rROL |
Free enzyme |
BR |
No |
1st |
Y: 64.2% |
[72] |
|
|
Triolein + MeOH |
rROL |
IA onto RelyZymeTM OD403S |
BR |
No |
1st |
Y: 82.6% |
[72] |
|
|
Triolein + EtOH |
rROL |
IA onto RelyZymeTM OD403S |
BR |
No |
1st |
Y:100.7% |
[72] |
|
|
JO + MeOH |
rROL |
IA onto Lewatit VP OC 1600 |
BR |
Yes |
2nd |
Y: 61% OS: after 10 cycles Y decreased a 40% |
[152] |
|
|
JO + MeOH |
rROL |
IA onto LifetechTM ECR1030M |
BR |
Yes |
2nd |
Y: 63% OS: after 10 cycles Y decreased a 40% |
[152] |
|
|
JO + MeOH |
rROL |
IA onto LifetechTM AP1090M |
BR |
Yes |
2nd |
Y: 55% OS: after 10 cycles Y decreased a 25% |
[152] |
|
|
JO + MeOH |
rROL |
CI onto LifetechTM ECR8285M |
BR |
Yes |
2nd |
Y: 63% OS: after 10 cycles Y decreased a 60% |
[152] |
|
|
JO + MeOH |
rROL |
CI onto Amberlita IRA 96 |
BR |
Yes |
2nd |
Y: 68% OS: after 10 cycles Y decreased a 20% |
[152] |
|
|
OO + MeOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 77% |
[153] |
|
|
OO + EtOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 62% |
[153] |
|
|
OO + Propanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 46% |
[153] |
|
|
OO + Butanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 18% |
[153] |
|
|
SYO + MeOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 50% |
[153] |
|
|
SYO + EtOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 46% |
[153] |
|
|
SYO + Propanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 35% |
[153] |
|
|
SYO + Butanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 10% |
[153] |
|
|
CO + MeOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 70% |
[153] |
|
|
CO + EtOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 56% |
[153] |
|
|
CO + Propanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 43% |
[153] |
|
|
CO + Butanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 16% |
[153] |
|
|
SO + MeOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 32% |
[153] |
|
|
SO + EtOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 28% |
[153] |
|
|
SO + Propanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 17% |
[153] |
|
|
SO + Butanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
1st |
Y: 7% |
[153] |
|
|
Algal oil + MeOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
3rd |
Y: 63% |
[153] |
|
|
Algal oil + EtOH |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
3rd |
Y: 55% |
[153] |
|
|
Algal oil + Propanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
3rd |
Y: 40% |
[153] |
|
|
Algal oil + Butanol |
prorROL |
IA onto Amberlite XAD 761 |
BR |
No |
3rd |
Y: 13% |
[153] |
|
|
ALO + MeOH |
rROL |
CI onto AP and GA treated ReliZymeTM HFA403 |
BR |
Yes |
2nd |
Y: 28.62% OS: after 9 cycles, Y decreased a 43% |
[104] |
|
|
JO + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
Yes |
2nd |
Y: 88.6% OS: after 6 cycles Y decreased a 21% |
[154] |
|
|
OA + MeOH |
proROL |
WCB immobilised into BSPs |
BR |
No |
1st |
Y: 80% OS: after 8 cycles, almost no activity loss. |
[155] |
|
|
Rice bran oil + MeOH |
proROL |
IA onto rod-like mesoporous silica |
BR |
No |
1st |
Y: 81.7% OS: after 3 cycles Y decreased to 67.7% |
[156] |
|
|
JO + MeOH |
proROL |
IE into polyvinyl alcohol—alginate matrix |
BR |
No |
2nd |
Yield: 87.1% |
[157] |
|
|
ALO + MeOH |
rROL |
IA Octadecyl-Sepabeads |
BR |
Yes |
2nd |
Y: 58.31% OS: after 2 cycles Y decreased to 54.67% |
[158] |
|
|
Tung oil + MeOH |
proROL |
CI onto Amberlite IRA 93 |
BR |
Yes |
2nd |
Y: 91.9% OS: after 6 cycles Y decreased to 85.1% |
[32] |
|
|
Babassu oil + EtOH |
proROL |
WCB immobilised into BSPs |
BR |
No |
1st |
Y: 74.15% |
[159] |
|
Regarding third-generation biodiesel, microalgae and waste oils have been the most studied substrates. The former has several advantages that make the overall process of biodiesel production more environmentally friendly as microalgae oil production involves atmospheric CO2 fixation and can use domestic wastewater like growth substrate facilitating its posterior treatment. However, the main drawbacks for microalgae oil employment are the scale-up of photobioreactors and lipids extraction [160][161]. Nevertheless, ROL has been satisfactorily employed with this substrate, for instance, with oils extracted from Nannochloropsis gaditana [132][133][134], Botryococcus braunii [134] and Chlorella vulgaris [135]. Actually, with the last one, fatty acid methyl esters (FAME) conversions over 70% were obtained indicating ROL suitability for biodiesel production with microalgae oil. Additionally, oils extracted from oleaginous yeasts, such as Candida sp. LEB-M3, have been also employed. The use of yeasts becomes important in biodiesel refineries as they might grow in the glycerol coming from this biofuel production [143]. Regarding waste oils, they have a significant potential in biodiesel industry because of their relevance in circular economy strategies, which aim to avoid residue generation by seeking new applications to waste [162][163]. Moreover, considering the tight economic competition between biodiesel and fossil fuels, cheap raw materials are required. In fact, the cost of the feedstocks is more than the 70% of the total cost of biodiesel. Thus, oleaginous wastes might help lowering these percentage and making enzymatic biodiesel production feasible [164]. Sludge from palm oil [136] and spent coffee grounds [137]can be found amongst some of the oleaginous residues studied in biodiesel production with ROL. However, waste cooking oil is the foremost substrate of this category because it is inexpensive and, through its employment in biodiesel synthesis, public institutions avoid the great cost of its management [165][166]. Relevant results have been published with WCO, for instance, Bharathiraja et al. [138] reported maximum triglyceride conversion of 94%. Nevertheless, not many studies dealing with ROL and WCO have been published and, considering the great relevance of this substrate for biodiesel industry, it could be a possible research target for future projects.
Biocatalysts operational stability, reusability and price are related and essential traits that must be considered in enzymatic biodiesel production because of the high cost of enzymes and the tight economic competition with conventional diesel. Some approaches have focused on cutting prices of the enzymes through heterologous production, as it has been explained in the previous section. Besides, other strategies have centred on lipase immobilisation. This technique allows enzyme reutilisation and generally enhances enzyme stability [167][168][169]. In the following paragraphs, the different immobilisation strategies assessed with ROL in biodiesel production will be introduced.
Earlier attempts of employing this enzyme in biodiesel synthesis were principally based on whole-cell biocatalysts (WCBs). Thus, the enzyme acts confined in its natural cellular environment, which protects the lipase from inactivation and degradation. Moreover, as no downstream processes of the biocatalyst are needed, its final cost is considerably lowered [170]. Syed et al. [139] immobilised lipase-producing R. oryzae cells into alginate beads and employed them in biodiesel production with jatropha and karanja oil. A response surface optimisation was applied and under the best conditions, biodiesel yields of 73.5% and 72.5% with each respective oil were obtained. In addition, operational stability of the biocatalyst was evaluated and after six cycles, just an activity loss of 20% was reported. Even if free cells, without immobilisation into alginate beads, could have been used in biodiesel production, Sun et al. [140] stated the suitability of cell immobilisation to avoid enzyme leakage and denaturation. This author immobilised R. oryzae fungus cells onto biomass support particles (BSPs) and obtained higher operational stability than using free cells. Moreover, to further minimise the enzyme leakage and deactivation, the crosslinking agent glutaraldehyde was used for immobilised cells treatment. The crosslinked biocatalyst obtained better FAMEs yields and operational stability. In the same sense, glutaraldehyde treatment of WCBs—also called WCBs stabilisation—was reported by Ban et al. [141] as well. Lately, He et al. [122] employed this strategy too and obtained a ROL biocatalyst with increased operational stability. After six reactions cycles, more than 90% of initial activity was maintained. However, WCBs show higher complexity in being reused and worse conversion rates than free lipases immobilised onto acrylic resins [170]. For instance, Bharathiraja et al. [138] published that WCBs exhibit worse reaction rate than immobilised purified proROL because of diffusional problems. Therefore, considering these inconveniences and how heterologous production of ROL has been improved, the use of free ROL and its subsequent immobilisation have gained importance amongst the published works.
Traditionally, lipases have been immobilised through adsorption, particularly onto hydrophobic supports—generally acrylic resins with hydrophobic superficial groups such as octadecyl or divinylbenzene—because of the presence of a large hydrophobic patch around the catalytic triad of the lipases that enables an easy immobilisation and might lead to their hyperactivation [171][172]. However, during biodiesel enzymatic synthesis, highly non-polar reaction mediums are employed that might cause enzyme desorption and in consequence, poor biocatalyst operational stability [51]. Nevertheless, some authors have used ROL with this immobilisation technique and obtained outstanding stability results. For instance, Bonet-Ragel et al. [142] reported that after six consecutive reaction cycles, the biocatalysts retained more than the 60% of the initial activity, in accordance with the results published by Duarte et al. [143] and Su et al. [144]. Moreover, in order to overcome the potential enzyme leakage when adsorption techniques are used, some published works have treated the obtained biocatalysts with crosslinking agents like glutaraldehyde, as it was previously explained for WCBs [130][173]. Notwithstanding these mentioned works and other listed in Table 4, ROL entrapment and covalent immobilisation are the most common immobilisation techniques. The former has been used not only with free ROL but with WCBs because it is an easy, fast and cheap immobilisation technique [174]. The most common entrapment strategies are based on polyvinyl alcohol and alginate employment [145][146][157][175]. Muanruksa et al. [136] obtained outstanding results with free proROL immobilised into alginate-polyvinyl alcohol beads. Esterification degrees over 90% were reported and the biocatalyst exhibited a high operational stability, 15 reaction cycles were done with almost no loss of activity.
Regarding covalent immobilisation, since the binding forces between the lipase and the supports are strong, obtained biocatalysts tend to show high stability, high resistance to extreme pH and temperature conditions and almost no enzyme leakage. However, these strong links between the enzyme and the support, as well as the harsh conditions employed during immobilisation process, might have a negative impact on the enzyme activity [176][177]. In any case, there are several studies that employ this immobilisation technique in biodiesel synthesis. Nematian et al. [147] immobilised proROL onto a superparamagnetic nanostructure and described that amongst the three different biocatalysts studied—two based on lipase-support electrostatic interactions and the third one on covalent-linkage—the covalently immobilised proROL showed higher conversion and operational stability. Bonet-Ragel et al. [115] covalently immobilised rROL onto glutaraldehyde pre-treated epoxide acrylic resins and studied its reaction performance and operational stability in biodiesel synthesis with methanol and ethanol as acyl-acceptors. Under the best conditions, yields close to the theoretical 100% were obtained after 360 min for methanol and 260 min for ethanol. In addition, regarding operational stability, no significant activity loss was observed after five consecutive reaction cycles with both alcohols. Besides, Luna et al. [126]described similar operational stability results with ethanol and sunflower oil as substrates, indicating that covalent immobilisation is an adequate technique for biodiesel synthesis with ROL.
In terms of operational strategy in biodiesel synthesis, although ROL has been described as a suitable industrial and solvent-tolerant enzyme, some improvements have been reported to obtain better reaction yields, higher stability or enhance the scale-up of the bioprocess. One of the most commonly employed approach is based on the stepwise addition of the alcohol as the interaction between the lipase and the alcohol is the main enzyme-deactivating factor [178][179]. Several authors have published works in which ROL and stepwise addition strategy have been employed [115][148]. Additionally, other authors have focused on seeking the most adequate acyl-acceptor—the one that has fewer negative effect on the enzyme—by testing different alcohols [138][175] and even the short-esters of the corresponding alcohols performing interesterification reactions [146][149]. Besides, regarding solvents employment, their absence in solvent-free systems has aroused as an interesting operational alternative because of the minimisation of biodiesel downstream processes and the avoidance of hazardous solvents, making the overall biotransformations more cost-effective and environmentally friendly [104][142][149][152].
Lately, the joint employment of both 1,3-regiospecific and non-specific lipases have been researched in order to accelerate biodiesel reaction rates and obtain higher yields [121]. Lee et al. [150] reported yields close to 100% in 2-h reaction and outstanding operational stabilities when using proROL and Candida rugosa lipase (CRL). Actually, the conversion yield was still 85% after 20 reaction cycles. In line with these results, Zeng et al. [151] described higher biodiesel production rates when employing together proROL and CRL.
Regarding the scale-up of biodiesel production using ROL, Canet et al. [119] compared packed bed reactor (PBR) with stirred tank reactor (STR) in biodiesel synthesis with rROL immobilised through hydrophobic adsorption. Results showed a higher reaction rate with STR than PBR but, just the opposite outcome when operational stability was the analysed trait. Other authors have also employed PBRs [125][131][145] or even more genuine reactors such as the magnetically stabilised fluidised bed reactor [124] or three-phase bioreactors [120]. However, there are not many works related to the scale-up of biodiesel production with ROL considering the vast amount of research papers published dealing with this biocatalyst. Therefore, more research in this field could be relevant for future projects.
4.2. Structured Lipids Production
Fats and oils are consumed in daily diets as an important source of energy, essential fatty acids and fat-soluble nutrients. Their functional, nutritional and organoleptic properties depend on their composition in saturated and polyunsaturated fatty acids, fatty acid chain length and on the distribution of the different fatty acids in the triacylglycerols (TAGs) (position sn-1, sn-3 or sn-2). Therefore, by modifying the fatty acids composition or its profile, lipids with improved properties might be obtained, the so-called structured lipids (SL). Currently, there are various SLs of commercial interest whose properties have been widely described (Table 5), (i) low caloric and dietetic triacylglycerols that include TAGs with medium-chains (MMM) and TAGs with short- and medium-chain fatty acids in sn-1 and sn-3 and a long-chain fatty acids in sn-2 position, SLS and MLM respectively; (ii) human milk fat substitutes (HMFS), (iii) cocoa butter equivalents (CBE), (iv) trans- free plastic fats, (v) triacylglycerols rich in specific long-chain and polyunsaturated fatty acids (PUFAs) and recently, even (vi) diacylglycerols (DAG) and monoacylglycerols (MAG) have been considered as SLs [180][181].
Table 5. Definition and properties of the main commercially relevant structured lipids.
|
SL Type |
Definition |
Properties |
Ref. |
|
Low caloric and dietetic TAGs |
Present lower caloric value than conventional oils and fats. SLS-, MLM- and MMM- type TAGs. |
M and S fatty acids present lower caloric value than their counterparts L. M fatty acids have lower tendency to get accumulated. Released M fatty acids can be directly absorbed and provide readily energy in the liver. |
|
|
Human milk fat substitutes (HMFS) |
Mimic the fatty acid profile of human milk. Contain oleic (30–35%), palmitic (20–30%), linoleic (7–14%) and stearic acids (5.7–8%). Palmitic acid mainly in sn-2 position. |
Promote palmitic acid absorption as 2-monoacylpalmitate Promote calcium absorption |
|
|
Cocoa butter equivalents (CBE) |
Mimic the scarce natural cocoa butter Mainly formed by saturated fatty acids (stearic and palmitic acids) in sn-1,3 and monounsaturated fatty acids (oleic acid) in sn-2 position. |
Desirable polymorph is β form Similar organoleptic properties to cocoa butter |
|
|
Trans-free plastic fats |
Mimic trans fatty acids containing hydrogenated vegetable oils. |
Avoid potential cardiovascular diseases caused by trans fatty acids. |
|
|
TAGs rich in specific long-chain and polyunsaturated fatty acids (PUFAs) |
Modified TAGs containing a combination of n-3 and n-6 PUFAs to enhance nutritional values. Mainly eicosapentaenoic (EPA) and docosahexaenoic acid (DHA) are employed. |
EPA decreases blood viscosity, platelets aggregation and promotes vasodilation. DHA promotes sensorial and neuronal maturation in babies. |
|
|
MAGs and DAGs |
Modified lipids containing one or two fatty acids linked to a glycerol |
Non-ionic surfactants capable of using as emulsifiers in the food industry. 1,3-DAGs reduce serum TAGs level and supress body fat accumulation |
SLs production can be carried out through chemical or enzymatical processes, the latter having several advantages when compared to chemical catalysis [192]. Hence, in the same way as stated for biodiesel synthesis in the previous section, enzymatically catalysed reactions allow milder reaction conditions what in this case, as well as lowering energy consumption, might lead to a reduction in the loss of original attributes of temperature-sensitive substrates and products. Moreover, through enzymatic catalysis, the use of deleterious solvents can be avoided enabling a safer and more environmentally friendly food production. However, the most remarkable advantage of lipase employment in this biotransformation is their specificity and selectivity [193][194]. Concretely, 1,3-regiospecific lipases like ROL arouse a keen interest because of their capacity to only modify the sn-1 and sn-3 positions of TAGs—even though acyl-migration phenomena might occur depending on reaction conditions.
Table 6 shows a summary of the latest published works about SLs synthesis employing ROL. Nunes et al. [195] produced MLM-type SLs by acidolysis of olive oil with capric and caprylic acids. The employed biocatalysts were rROL produced in K. phaffii and commercial native ROL (proROL), both of them covalently immobilised onto Eupergit© C and modified Sepiolite. Noticeably, rROL showed a better performance than the native lipase, the percentages of incorporated capric and caprylic acids were higher as well as the operational stability. In spite of the use of pure or commercial substrates, oleaginous wastes or even non-commercially profitable oils might also be employed for MLM-type SLs synthesis with ROL. For instance, Mota et al. [196] described how low-calorie SLs of MLM-type can be produced using oil extracted from spent coffee grounds and oil from olive pomace with proROL immobilised onto magnetic nanoparticles. In the same line, Costa et al. [197] synthesised MLM-type SLs with the oil extracted from grapeseeds of Vitis vinifera L., which are a by-product of the wine industry. Moreover, instead of residual oils, Nagao et al. [198] employed the oil from the oleaginous microorganism Mortierella alpina to produce MLMs rich in arachidonic acid, a precursor of several hormones.
Table 6. Summary of structured lipids production with Rhizopus oryzae lipase as main biotcatalyst.
|
Product |
Substrates |
Reaction Type |
Lipase |
Immobilisation Technique |
ID/OS |
Ref. |
|
MLM |
OO + CRA |
Acidolysis |
proROL/rROL |
CI onto Eupergit®C/sepiolite (AlPO4-sepiolite) |
ID: 21.6%. OS: half-life 159 h |
[195] |
|
MLM |
OO + CA |
Acidolysis |
proROL/rROL |
CI onto Eupergit®C/sepiolite (AlPO4-sepiolite) |
ID: 34.82%. OS: half-life 136 h |
[195] |
|
MLM |
SCG + CA |
Acidolysis |
proROL |
CI onto GA treated MNP |
ID: 50% |
[196] |
|
MLM |
SCG + ethyl caprate |
Interesterification |
proROL |
CI onto GA treated MNP |
ID: 26% |
[196] |
|
MLM |
OP + CA |
Acidolysis |
proROL |
CI onto GA treated MNP |
ID: 51% OS: 6.8 batches |
[196] |
|
MLM |
OP + ethyl caprate |
Interesterification |
proROL |
CI onto GA treated MNP |
ID: 46%. OS: 9.1 batches |
[196] |
|
MLM |
Grapeseed oil + CRA |
Acidolysis |
rROL |
CI onto Amberlite IRA 96 |
ID: 54%. OS: half-life 166 h |
[197] |
|
MLM |
Grapeseed oil + CA |
Acidolysis |
rROL |
CI onto Amberlite IRA 96 |
ID: 69% OS: half-life 118 h |
[197] |
|
MLM |
TGA58F + CA |
Acidolysis |
proROL |
IA onto Dowex WBA |
ID: 64.6% |
[198] |
|
MLM |
TGA40 + CA |
Acidolysis |
proROL |
IA onto Dowex WBA |
ID: 62.8% |
[198] |
|
MLM |
TGA55E + CA |
Acidolysis |
proROL |
IA onto Dowex WBA |
ID: 64.8% OS: 90 days in PBR1 dropped 10% |
[198] |
|
MLM |
OO + CRA |
Acidolysis |
rROL |
CI onto Eupergit® C/IA onto Lewatit VP OC 1600 |
OS: half time 2.4 batches (54.3 h) with Eupergit®C |
[199] |
|
MLM |
OO + CA |
Acidolysis |
rROL |
CI onto Eupergit® C/IA onto Lewatit VP OC 1600 |
OS: half time 10.2 batches (234 h) with Lewatit VP OC 1600 |
[199] |
|
MLM |
OO + CRA |
Acidolysis |
rROL |
CI onto Eupergit® C |
ID: 15.5% |
[200] |
|
MLM |
OO + CA |
Acidolysis |
rROL |
CI onto Eupergit® C |
ID: 33.3% |
[200] |
|
MLM |
OO + CRA |
Acidolysis |
rROL |
CI onto Amberlite IRA 96 |
ID: 76.9 |
[201] |
|
MLM |
OO + CA |
Acidolysis |
rROL |
CI onto Amberlite IRA 96 |
ID: 85.6% |
[201] |
|
HMFS |
PA enriched TAGs + OA enriched mixtures |
Acidolysis |
proROL |
IA onto Accurel® MP-1000 |
ID: OA in sn-1,3 67.2% - PA in sn-2 67.8%. OS: no activity loss in 10 uses (190 h) |
[202] |
|
HMFS |
Lard + FFA from EPAX 1050TG |
Acidolysis |
rROL |
CI onto Accurel® MP-1000 |
ID: 24 mol%. OS: after 4 batches, 55% of original activity |
[203] |
|
HMFS |
Tripalmitin + FFA from camelina oil |
Acidolysis |
rROL |
AI onto RelizymeTM OD403/S/CI onto Lewatit VP OC 1600 |
ID: 52% |
[204] |
|
TAGs rich in PUFAs |
cod liver + tuna oil + ethanol. |
Alcoholysis |
proROL |
IA onto Accurel® MP-1000 |
Alcoholysis ID: 72% OS: after 6 cycles, complete deactivation. |
[205] |
|
2-MAG from alcoholysis + CRA |
Esterification |
proROL |
IA onto Accurel® MP-1000 |
ID: 95%. OS: after 5 cycles, no activity loss. |
[205] |
|
|
TAGs rich in PUFAs |
Tuna oil + CRA |
Acidolysis |
proROL |
IA onto Accurel® MP-1000 |
OS: over one week |
[206] |
|
TAGs rich in PUFAs |
cod liver oil + ethanol 96% |
Alcoholysis |
proROL |
IA onto Accurel® MP-1000 |
Alcoholysis Y: 78%. OS: after 3 cycles, a 57% decrease |
[207] |
|
cod liver oil + 1-butanol |
Alcoholysis |
proROL |
IA onto Accurel® MP-1000 |
Alcoholysis Y: 78%. OS: after 3 cycles, no activity decrease |
[207] |
|
|
Esterification: 2-MAG from alcoholysis + CRA |
Esterification |
proROL |
IA onto Accurel® MP-1000 |
Esterification Y: 71%. |
[207] |
|
|
TAGs rich in PUFAs |
Fish oil + CRA |
Acidolysis |
proROL |
Non-immobilised |
ID: 2.5% |
[208] |
|
HMFS |
Milkfat + SYO |
Interesterification |
proROL |
EI into polysiloxane-PVA |
ID: 8.14%. OS: after 10 batches, no activity loss |
[209] |
|
CBE |
SO + SA-PA mixtures |
Acidolysis |
proROL |
IA onto Accurel® MP-1000 |
|
[210] |
Regarding HMFS, Esteban et al. [202] used several commercial lipases, amongst them proROL immobilised onto Accurel® MP-1000, to produce a TAG rich in palmitic acid in sn-2 and oleic acid in sn-1,3; the so called OPO, which is the main component of human milk TAGs. proROL showed the best performance in oleic acid incorporation and exhibited a high operational stability, after ten reuse cycles almost no activity loss was found. Simões et al. [203] also tested different lipases for HMFS production and reported that rROL immobilised onto Accurel® MP-1000 showed a similar performance to Novozymes 435 and Lipozyme RM IM in acidolysis reaction between lard and FFA mixture from fish oil rich in docosahexaenoic acid. Besides, Faustino et al. [204] immobilised rROL produced in K. phaffii onto two different supports, Lewatit VP OC 1600 and Relizyme OD403/S, and applied the formed biocatalysts in the production of HMFS rich in polyunsaturated fatty acids (PUFAs). The acidolysis reaction was carried out in solvent-free system between tripalmitin and FFAs (mainly linoleic and linolenic acids) from camelina oil, which proved to be a good source of PUFAs. According to the authors, the results obtained with rROL immobilised onto Lewatit VP OC 1600 were comparable to the commonly used commercial lipase Lipozyme RM IM.
Triacylglycerols rich in long-chain and polyunsaturated fatty acids have also been produced with ROL. In most of the cases, these SLs’ production is based on a two-step process in order to minimise the acyl migration phenomena [211]. In the first step, through alcoholysis reaction, 2-monoacylglycerols (2-MAGs) are obtained from oils containing TAGs rich in PUFAs or long-chain fatty acids in the mentioned sn-2 position, usually fish oils. Then, these 2-MAGs are esterified with other relevant FFA to obtain the nutritionally interesting TAGs rich in PUFAs. For instance, Muñio et al. [205] studied the performance of different commercial lipases, including proROL immobilised onto Accurel® MP-1000, in the process of alcoholysis of tuna and cod oil to obtain 2-MAGs and then, carry out their subsequent esterification with capric acid. In alcoholysis reaction the commercial lipase Novozyme 435 showed a better operational stability than Lipase D (commercial proROL), although the latter exhibited higher reaction yield. During esterification reaction, Lipase D obtained the highest SLs percentage (over 90%) in the mixture. Moreover, no loss in proROL activity was observed after at least five reaction cycles. Hita et al. [206] and Rodriguez et al. [207] reported similar results with immobilised proROL.
With respect to CBE, although Ray et al. [210] described the kinetics of the acidolysis of high oleic sunflower oil with stearic–palmitic acid mixtures that, after further fractionation of the product, could be potentially used in CBE formulations, ROL has not been extensively used for CBE production. Therefore, this subject might be a great research target for future projects, as well as DAG and MAG synthesis, which have not been specifically treated but just as a minor topic during other products synthesis, like biodiesel.
4.3. Flavour Esters Production
Flavour and aromatic esters are widely found in nature and have pleasant organoleptic attributes, including fruity, floral, spicy, creamy or nutty aromas. These traits made them suitable as ingredients for food, beverages, cosmetics, pharmaceuticals, chemicals and personal care products, like perfumes, body lotions, shampoos and other toiletries [212][213]. In general, most of the flavour and fragrance compounds are produced through extraction from their natural source, usually fruits, plants and flowers. However, they are found in the environment in low concentrations making the extraction a costly process and not viable to fulfil their growing demand. Therefore, chemical and enzymatic synthesis procedures have aroused to solve flavour esters scarcity [214][215]. Noticeably, the latter exhibits a significant advantage—notwithstanding the already explained benefits of enzymatic synthesis over chemical one in the previous sections—which is the capacity to label the obtained products as natural according to European Legislation (EC 1334/2008) if and when the employed reactants are also natural. Thereby, the use of enzymes satisfies consumers trend towards natural products and boosts economic value of the obtained flavour esters [212]. In fact, as well as ROL, other lipases have been used for flavour esters production, for instance, the commercial Novozym® 435 (Candida antartica lipase B) [214][215], Candida rugosa lipase [216][217] and Burkholderia cepacia lipase [218].
Ethyl butyrate is an important component of many fruit flavours such as pineapple, passion fruit and strawberry [219]. The enzymatic synthesis of this compound can be carried out through esterification of butyric acid and ethanol. Guillen et al. [220] immobilised rROL onto three different supports, EP100, Eupergit®CM and Octadecyl-Sepabeads to test them in this esterification reaction. In terms of reaction rate and yield, rROL immobilised onto EP100 showed the best performance. However, rROL immobilised onto Octadecyl-Sepabeads exhibited the highest operational stability. Consequently, this biocatalyst was used for further research in which the effects of butyric acid and ethanol concentration were studied through DoE strategy to maximise the reaction rate and final yield [221]. The obtained results indicated that the suitable acid:alcohol ratio for maximum yield was 1.45 and that the higher the butyric acid concentration the higher the reaction rate. However, as previously described by Grosso et al. [222], elevated concentrations of butyric acid led to enzyme deactivation.
Butyl acetate is another flavour ester with resembling organoleptic properties to pineapple flavour whose production with ROL was reported by Ben Salah et al. [223]. The synthesis of this compound was carried out through esterification reaction of butanol and acetic acid with immobilised proROL onto Celite 545—as preliminary results of the reaction with free enzyme showed poor yield and they were clearly exceeded by the immobilised biocatalyst. According to the authors, solvent-free reaction was chosen as the most suitable strategy because of the easier product purification and lower toxicity and inflammability. In these conditions, a maximum yield of 60% was obtained and the biocatalyst was stable for three consecutive cycles without a decrease in synthesis activity.
Besides esterification, transesterification reaction catalysed by ROL has also been employed for flavour esters synthesis, for example, Kumari et al. [224] reported isoamyl acetate ester synthesis—pleasant banana flavour—through isoamyl alcohol and vinyl acetate transesterification with immobilised proROL. Furthermore, as stated by these authors, the inhibitory effect of the acid [225] was avoided through the use of transesterification reaction with vinyl acetate ester instead of esterification reaction with the corresponding acid. Under optimal conditions, a conversion of 95% in 8 h of reaction was obtained including a great operational stability, after three reaction cycles no activity loss was detected. Garlapati et al. [226] described the use of covalently immobilised proROL onto activated silica to produce through transesterification reactions methyl butyrate and octyl acetate, flavour esters with pineapple and orange odours respectively. As a result of an optimisation process, authors reached high reaction yields in solvent-free system, 70.42% in 14 h and 92.35% in 12 h for methyl butyrate and octyl acetate respectively. Moreover, in both cases, the biocatalyst was reusable for five times retaining a relative activity of more than 95%. Transesterification reaction was as well employed for citronellol esters synthesis with immobilised proROL into HPMC–PVA polymer (hydroxypropyl methyl cellulose—polyvinyl alcohol) and in supercritical carbon dioxide reaction medium [227]. For the three studied flavour esters (citronellol acetate, citronellol butyrate and citronellol laurate) final yields over 90% were achieved indicating the suitability of this biocatalysts and the proposed system for these biotransformations.
4.4. Resolution of Racemic Mixtures
Enantiomerically pure compounds are very attractive for the preparation of a wide range of products, particularly in food and pharmaceutical industries where the desired organoleptic properties or effects might be only related to one of the isomers. Therefore, racemic resolution processes become relevant and arouse the interest in lipases considering the enantioselectivity and specificity of these enzymes [228][229].
Palomo et al. [230] employed proROL to carry out the enzymatic resolution of (R)-glycidyl butyrate because of its importance in linezolid synthesis. This product is already sold as a treatment for multidrug resistant Gram-positive infections. According to these authors, they followed the ‘conformational engineering’ strategy, that is, different techniques for proROL immobilisation were employed. This way, the enzyme structure would have different rigidity or the microenvironment surrounding the enzyme would alter the exact shape of the open form of the lipase influencing its catalytic performance. Amongst the three different biocatalysts formed, the best enantiomeric excess (ee) was obtained with proROL immobilised through adsorption on dextran sulphate-coated sepabeads, 99% ee with a 55% conversion.
Benzoin is a relevant α-hydroxy ketone that might act as building block in organic synthesis. Songür et al. [231] described its enantioselective production from benzoin acetate through the employment of R. oryzae cell homogenates. The objective of using cell homogenates was to combine the enantioselective hydrolysis of proROL with the racemisation process of the racemase of R. oryzae in order to increase the ee and conversion values. This way, a final conversion of (S)-benzoin close to the 100% and 96% ee was achieved.
Covalently immobilised proROL onto Lewatit-aldehyde support has been reported as an adequate biocatalyst for asymmetric hydrolysis of dimethyl 3-phenylglutarate [232]. Under the best conditions, it was possible to obtain the (R)-methyl-3-phenylglutarate with a 92% ee and an yield in monoester of 97%.
(S)-enantiomer of ibuprofen is 160 more active than its (R)-enantiomer, which can even cause side effects in the gastrointestinal tract. Therefore, obtaining the adequate enantiomer becomes crucial in this case. Yousefi et al. [233] reported the use of immobilised proROL onto octadecyl sepharose to carry out the enantioselective resolution of racemic ibuprofens esters.
The racemic resolution of (R,S)-1-phenylethanol to produce (S)-1-phenylethanol, a chiral building block, was carried out with proROL-displaying yeast whole cell biocatalyst, that is, a S. cerevisiae strain genetically modified to display proROL on the cell surface. After 36 h of reaction, significant results were obtained, 97.3% yield and 93.3% ee [234]. The same biocatalyst was employed to catalyse the optical resolution of the pharmaceutical precursor (R,S)-1-benzyloxy-3-chloro-2-propyl monosuccinate. In this case, the operational stability of the biocatalysts was assessed and it was stable after at least eight reaction cycles [235].
Abbreviations
|
28proROL-gene |
Gene encoding a truncated prosequence of Rhizopus oryzae lipase 28 C-terminal amino acids fused to the N-terminal of the mature lipase region |
|
2-MAG |
2-monoacylglycerol |
|
ALO |
Alperujo oil |
|
BR |
Batch Reactor |
|
C |
TAG or FFA conversion (%) |
|
CA |
Capric acid |
|
CBE |
Cocoa butter equivalents |
|
CI |
Covalently immobilised or stabilised biocatalyst through crosslinking |
|
CO |
Canola oil |
|
CRA |
Caprylic acid |
|
CRL |
Candida rugosa lipase |
|
DAG |
Diacylglycerol |
|
DoE |
Design of experiments |
|
EDTA |
Ethylenediaminetetraacetic acid |
|
ee |
Enantiomeric excess |
|
entire-proROL |
Rhizopus oryzae lipase including the whole prosequence and mature sequence |
|
EPAX 1050TG |
TAG rich in omega-3 PUFAs |
|
EtOH |
Ethanol |
|
FAME |
Fatty acid methyl esters |
|
FFA |
Free fatty acid |
|
HMFS |
Human milk fat substitutes |
|
IA |
Immobilisation through adsorption |
|
ICL |
Isocitrate lyase |
|
ID |
Incorporation degree (%) |
|
IE |
Immobilisation through physical entrapment |
|
JO |
Jatropha oil |
|
KO |
Karanja oil |
|
L |
Long-chain fatty acid |
|
M |
Medium-chain fatty acid |
|
MAG |
Monoacylglycerol |
|
MeOH |
Methanol |
|
MSFBR |
Magnetically-stabilised fluidised bed reactor |
|
Mut+ |
Methanol utilisation plus phenotype |
|
Muts |
Methanol utilisation slow phenotype |
|
MW |
Molecular weight (kDa) |
|
NBS |
N-Bromosuccinimide |
|
OA |
Oleic acid |
|
OO |
Olive oil |
|
OP |
Olive pomace |
|
OPO |
TAG with oleic acid in sn-1,3 positions and palmitic acid in sn-2 position. |
|
OS |
Operational stability |
|
PA |
Palmitic acid |
|
PAOX |
Inducible Alcohol oxidase promoter |
|
PBR |
Packed bed reactor |
|
PFL |
Pseudomonas fluorescens lipase |
|
PMSF |
Phenylmethylsulfonyl fluoride |
|
proROL |
R. oryzae lipase containing the N-terminal of mature sequence attached to 28 C-terminal amino acids of the prosequence |
|
proROL-gene |
Gene encoding the prosequence of 97 amino acids fused to the N-terminal of the mature lipase region of 269 amino acids |
|
PUFA |
Polyunsaturated fatty acids |
|
PVA |
Polyvinylalcohol |
|
RO |
Rapeseed oil |
|
ROL |
Rhizopus oryzae lipase |
|
rROL |
Rhizopus oryzae lipase containing mature sequence of R. oryzae lipase |
|
rROL-gene |
Gene encoding the mature lipase |
|
S |
Short-chain fatty acid |
|
SA |
Stearic acid |
|
SCG |
Spent coffee ground |
|
SGLB |
Solid gas liquid bioreactor |
|
SL |
structured lipid |
|
SLLB |
Solid liquid liquid bioreactor |
|
SO |
Sunflower oil |
|
STR |
Stirred tank reactor |
|
SYO |
Soybean oil |
|
TAGs |
Triacylglycerols |
|
TGA40 |
commercial oil |
|
TGA55E |
Hydrolysed TGA40 oil |
|
TGA58F |
Mortierella alpina single-cell oil |
|
TPB |
Three phase bioreactor |
|
UBC1 |
Ubiquitin-conjugating enzyme |
|
UPR |
5’ upstream region |
|
WCB |
Whole cells biocatalyst |
|
WCO |
Waste cooking oil |
|
Y |
Yield (%) |
References
- Liliana Londoño-Hernández; Cristina Ramírez-Toro; Hector A. Ruiz; Juan A. Ascacio-Valdés; Miguel A. Aguilar-Gonzalez; Raúl Rodríguez-Herrera; Cristóbal N. Aguilar; Rhizopus oryzae – Ancient microbial resource with importance in modern food industry. International Journal of Food Microbiology 2017, 257, 110-127, 10.1016/j.ijfoodmicro.2017.06.012.
- Joseph Sebastian; Krishnamoorthy Hegde; Pratik Kumar; Tarek Rouissi; Satinder Kaur Brar; Bioproduction of fumaric acid: an insight into microbial strain improvement strategies. Critical Reviews in Biotechnology 2019, 39, 817-834, 10.1080/07388551.2019.1620677.
- Olfa Benabda; Sana M’Hir; Mariam Kasmi; Wissem Mnif; Moktar Hamdi; Optimization of Protease and Amylase Production by Rhizopus oryzae Cultivated on Bread Waste Using Solid-State Fermentation. Journal of Chemistry 2019, 2019, 1-9, 10.1155/2019/3738181.
- Barnita Ghosh; Rina Rani Ray; Current Commercial Perspective of Rhizopus oryzae: A Review. Journal of Applied Sciences 2011, 11, 2470-2486, 10.3923/jas.2011.2470.2486.
- Xiao-Wei Yu; Yan Xu; Rong Xiao; Lipases from the genus Rhizopus : Characteristics, expression, protein engineering and application. Progress in Lipid Research 2016, 64, 57-68, 10.1016/j.plipres.2016.08.001.
- H.Dietmar Beer; John E.G. McCarthy; Uwe T. Bornscheuer; Rolf D. Schmid; Cloning, expression, characterization and role of the leader sequence of a lipase from Rhizopus oryzae. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1998, 1399, 173-180, 10.1016/s0167-4781(98)00104-3.
- Riadh Ben Salah; Habib Mosbah; Ahmed Fendri; Ali Gargouri; Youssef Gargouri; Hafedh Mejdoub; Biochemical and molecular characterization of a lipase produced by Rhizopus oryzae. FEMS Microbiology Letters 2006, 260, 241-248, 10.1111/j.1574-6968.2006.00323.x.
- Adel Sayari; Fakher Frikha; Nabil Miled; Hounaida Mtibaa; Yassine Ben Ali; Robert Verger; Youssef Gargouri; N-terminal peptide ofRhizopus oryzaelipase is important for its catalytic properties. FEBS Letters 2005, 579, 976-982, 10.1016/j.febslet.2004.12.068.
- U Derewenda; L Swenson; Y Wei; R Green; P M Kobos; R Joerger; M J Haas; Z S Derewenda; Conformational lability of lipases observed in the absence of an oil-water interface: crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar.. Journal of Lipid Research 1994, 35, 524-534, 10.2210/pdb1tic/pdb.
- Mitsutaka Kohno; Wataru Kugimiya; Yukio Hashimoto; Yuhei Morita; Purification, Characterization, and Crystallization of Two Types of Lipase fromRhizopus niveus. Bioscience, Biotechnology, and Biochemistry 1994, 58, 1007-1012, 10.1271/bbb.58.1007.
- Min Yang; Xiao-Wei Yu; Haiyan Zheng; Chong Sha; Caifeng Zhao; Meiqian Qian; Yan Xu; Role of N-linked glycosylation in the secretion and enzymatic properties of Rhizopus chinensis lipase expressed in Pichia pastoris.. Microbial Cell Factories 2015, 14, 40, 10.1186/s12934-015-0225-5.
- Xiao-Wei Yu; Min Yang; Chuanhuan Jiang; Xiaofeng Zhang; Yan Xu; N-Glycosylation Engineering to Improve the Constitutive Expression of Rhizopus oryzae Lipase in Komagataella phaffii. Journal of Agricultural and Food Chemistry 2017, 65, 6009-6015, 10.1021/acs.jafc.7b01884.
- Hans-Dietmar Beer; Gerd Wohlfahrt; Rolf D. Schmid; John E. G. McCarthy; The folding and activity of the extracellular lipase of Rhizopus oryzae are modulated by a prosequence. Biochemical Journal 1996, 319, 351-359, 10.1042/bj3190351.
- Yu-Jen Chen; Masayori Inouye; The intramolecular chaperone-mediated protein folding. Current Opinion in Structural Biology 2008, 18, 765-770, 10.1016/j.sbi.2008.10.005.
- Shouji Takahashi; Mitsuyoshi Ueda; Haruyuki Atomi; Hans D. Beer; Uwe T. Bornscheuer; Rolf D. Schmid; Atsuo Tanaka; Extracellular production of active Rhizopus oryzae lipase by Saccharomyces cerevisiae. Journal of Fermentation and Bioengineering 1998, 86, 164-168, 10.1016/s0922-338x(98)80055-x.
- Mitsuyoshi Ueda; Shouji Takahashi; Motohisa Washida; Seizaburo Shiraga; Atsuo Tanaka; Expression of Rhizopus oryzae lipase gene in Saccharomyces cerevisiae. Journal of Molecular Catalysis B: Enzymatic 2002, 17, 113-124, 10.1016/s1381-1177(02)00018-8.
- Wei-Ning Niu; Zhao-Peng Li; Tianwei Tan; Secretion of Pro- and Mature Rhizopus arrhizus Lipases by Pichia pastoris and Properties of the Proteins. Molecular Biotechnology 2006, 32, 073-082, 10.1385/mb:32:1:073.
- Jian-Rong Wang; Yang-Yuan Li; Shude Xu; Peng Li; Jing-Shan Liu; Dan-Ni Liu; High-Level Expression of Pro-Form Lipase from Rhizopus oryzae in Pichia pastoris and Its Purification and Characterization. International Journal of Molecular Sciences 2013, 15, 203-217, 10.3390/ijms15010203.
- S. Takahashi; M. Ueda; A. Tanaka; Function of the prosequence for in vivo folding and secretion of active Rhizopus oryzae lipase in Saccharomyces cerevisiae. Applied Microbiology and Biotechnology 2001, 55, 454-462, 10.1007/s002530000537.
- Abderaouf Ben Salah; Adel Sayari; Robert Verger; Youssef Gargouri; Kinetic studies of Rhizopus oryzae lipase using monomolecular film technique. Biochimie 2001, 83, 463-469, 10.1016/s0300-9084(01)01283-4.
- S. Takahashi; M. Ueda; A. Tanaka; Independent production of two molecular forms of a recombinant Rhizopus oryzae lipase by KEX2 -engineered strains of Saccharomyces cerevisiae. Applied Microbiology and Biotechnology 1999, 52, 534-540, 10.1007/s002530051556.
- Josu López-Fernández; Juan J. Barrero; Maria Dolors Benaiges; Francisco Valero; Truncated Prosequence of Rhizopus oryzae Lipase: Key Factor for Production Improvement and Biocatalyst Stability. Catalysts 2019, 9, 961, 10.3390/catal9110961.
- Shinji Hama; Sriappareddy Tamalampudi; Naoki Shindo; Takao Numata; Hideki Yamaji; Hideki Fukuda; Akihiko Kondo; Role of N-terminal 28-amino-acid region of Rhizopus oryzae lipase in directing proteins to secretory pathway of Aspergillus oryzae. Applied Microbiology and Biotechnology 2008, 79, 1009-18, 10.1007/s00253-008-1502-6.
- Stefan Minning; Claudia Schmidt-Dannert; Rolf D Schmid; Functional expression of Rhizopus oryzae lipase in Pichia pastoris: high-level production and some properties. Journal of Biotechnology 1998, 66, 147-156, 10.1016/s0168-1656(98)00142-4.
- Miklós Takó; Alexandra Kotogan; Tamás Papp; Shine Kadaikunnan; Naiyf S. Alharbi; Csaba Vágvölgyi; Purification and Properties of Extracellular Lipases with Transesterification Activity and 1,3-Regioselectivity from Rhizomucor miehei and Rhizopus oryzae. Journal of Microbiology and Biotechnology 2017, 27, 277-288, 10.4014/jmb.1608.08005.
- Abel Hiol; Marie D. Jonzo; Nathalie Rugani; Danielle Druet; Louis Sarda; Louis Claude Comeau; Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzyme and Microbial Technology 2000, 26, 421-430, 10.1016/s0141-0229(99)00173-8.
- Yuji Shimada; Mieko Iwai; Yoshio Tsujisaka; Reversibility of the Modification of Rhizopus delemar Lipase by Phosphatidylcholine1. The Journal of Biochemistry 1981, 89, 937-942, 10.1093/oxfordjournals.jbchem.a133277.
- Marina Guillén; Maria Dolors Benaiges; Francisco Valero; Comparison of the biochemical properties of a recombinant lipase extract from Rhizopus oryzae expressed in Pichia pastoris with a native extract. Biochemical Engineering Journal 2011, 54, 117-123, 10.1016/j.bej.2011.02.008.
- Kh. Pashangeh; M. Akhond; H.R. Karbalaei-Heidari; G. Absalan; Biochemical characterization and stability assessment of Rhizopus oryzae lipase covalently immobilized on amino-functionalized magnetic nanoparticles. International Journal of Biological Macromolecules 2017, 105, 300-307, 10.1016/j.ijbiomac.2017.07.035.
- Michael J. Haas; David J. Cichowicz; David G. Bailey; Purification and characterization of an extracellular lipase from the fungusRhizopus delemar. Lipids 1992, 27, 571-576, 10.1007/bf02536112.
- Majda Essamri; Valérie Deyris; Louis Comeau; Optimization of lipase production by Rhizopus oryzae and study on the stability of lipase activity in organic solvents. Journal of Biotechnology 1998, 60, 97-103, 10.1016/s0168-1656(97)00193-4.
- Xiao-Wei Yu; Chong Sha; Yong-Liang Guo; Rong Xiao; Yan Xu; High-level expression and characterization of a chimeric lipase from Rhizopus oryzae for biodiesel production. Biotechnology for Biofuels 2013, 6, 29-29, 10.1186/1754-6834-6-29.
- Riadh Ben Salah; Ali Gargouri; Robert Verger; Youssef Gargouri; Hafedh Mejdoub; Expression in Pichia pastoris X33 of His-tagged lipase from a novel strain of Rhizopus oryzae and its mutant Asn 134 His: purification and characterization. World Journal of Microbiology and Biotechnology 2009, 25, 1375-1384, 10.1007/s11274-009-0024-4.
- Jayshree B. Kantak; Asmita A Prabhune; Characterization of Smallest Active Monomeric Lipase from Novel Rhizopus Strain: Application in Transesterification. Applied Biochemistry and Biotechnology 2012, 166, 1769-1780, 10.1007/s12010-012-9584-0.
- Chun Li; Guofang Zhang; Ning Liu; Libo Liu; Preparation and Properties of Rhizopus oryzae Lipase Immobilized Using an Adsorption-Crosslinking Method. International Journal of Food Properties 2016, 19, 1776-1785, 10.1080/10942912.2015.1107732.
- C.N.A. Razak; A.B. Salleh; R. Musani; M.Y. Samad; M. Basri; Some characteristics of lipases from thermophilic fungi isolated from palm oil mill effluent. Journal of Molecular Catalysis B: Enzymatic 1997, 3, 153-159, 10.1016/s1381-1177(96)00035-5.
- Zhilin Li; Xun Li; Ye Wang; Youdong Wang; Fei Wang; Jianchun Jiang; Expression and characterization of recombinant Rhizopus oryzae lipase for enzymatic biodiesel production. Bioresource Technology 2011, 102, 9810-9813, 10.1016/j.biortech.2011.07.070.
- Xin Song; Xiaoyu Qi; Bin Hao; Yinbo Qu; Studies of substrate specificities of lipases from different sources. European Journal of Lipid Science and Technology 2008, 110, 1095-1101, 10.1002/ejlt.200800073.
- Rafael Matsumoto Pereira; Grazielle S. S. Andrade; Heizir Ferreira De Castro; Maria Gabriela Nogueira Campos; Performance of Chitosan/Glycerol Phosphate Hydrogel as a Support for Lipase Immobilization. Materials Research 2017, 20, 190-201, 10.1590/1980-5373-mr-2017-0091.
- Tigran V. Yuzbashev; Evgeniya Y. Yuzbasheva; Tatiana V. Vibornaya; Tatiana I. Sobolevskaya; I. A. Laptev; Alexey V. Gavrikov; Sergei P Sineoky; Production of recombinant Rhizopus oryzae lipase by the yeast Yarrowia lipolytica results in increased enzymatic thermostability. Protein Expression and Purification 2012, 82, 83-89, 10.1016/j.pep.2011.11.014.
- Sunita Adak; Rintu Banerjee; Sunita Adak Rintu Banerjee; Sunita Adak And Rintu Banerjee; Biochemical Characterisation of a Newly Isolated Low Molecular Weight Lipase from Rhizopus oryzae NRRL 3562. Enzyme Engineering 2013, 2, 118-25, 10.4172/2329-6674.1000118.
- Maha Karra-Châabouni; Ines Bouaziz; Sami Boufi; Ana Maria Botelho Do Rego; Youssef Gargouri; Physical immobilization of Rhizopus oryzae lipase onto cellulose substrate: Activity and stability studies. Colloids and Surfaces B: Biointerfaces 2008, 66, 168-177, 10.1016/j.colsurfb.2008.06.010.
- Joseph D. Schrag; Miroslaw Cygler; 1·8 Å Refined Structure of the Lipase from Geotrichum candidum. Journal of Molecular Biology 1993, 230, 575-591, 10.1006/jmbi.1993.1171.
- P Grochulski; Y Li; Joseph D Schrag; F Bouthillier; P Smith; D Harrison; B Rubin; M Cygler; Insights into interfacial activation from an open structure of Candida rugosa lipase.. Journal of Biological Chemistry 1993, 268, 12843-7, 10.2210/pdb1crl/pdb.
- M.E.M. Noble; A. Cleasby; L.N. Johnson; M.R. Egmond; L.G.J. Frenken; The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Letters 1993, 331, 123-128, 10.1016/0014-5793(93)80310-q.
- U. Derewenda; L. Swenson; R. Green; Y. Wei; G.G. Dodson; S. Yamaguchi; M.J. Haas; Z.S. Derewenda; An unusual buried polar cluster in a family of fungal lipases. Nature Structural & Molecular Biology 1994, 1, 36-47, 10.1038/nsb0194-36.
- Nipon Sarmah; D. Revathi; G. Sheelu; K. Yamuna Rani; S. Sridhar; V. Mehtab; C. Sumana; Recent advances on sources and industrial applications of lipases. Biotechnology Progress 2017, 34, 5-28, 10.1002/btpr.2581.
- Faez Iqbal Khan; Dongming Lan; Rabia Durrani; Weiqian Huan; Zexin Zhao; Yonghua Wang; The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Frontiers in Bioengineering and Biotechnology 2017, 5, 16, 10.3389/fbioe.2017.00016.
- Atsushi Satomura; Kouichi Kuroda; Mitsuyoshi Ueda; Generation of a Functionally Distinct Rhizopus oryzae Lipase through Protein Folding Memory. PLOS ONE 2015, 10, e0124545, 10.1371/journal.pone.0124545.
- Seizaburo Shiraga; Mitsuyoshi Ueda; Shouji Takahashi; Atsuo Tanaka; Construction of the combinatorial library of Rhizopus oryzae lipase mutated in the lid domain by displaying on yeast cell surface. Journal of Molecular Catalysis B: Enzymatic 2002, 17, 167-173, 10.1016/s1381-1177(02)00024-3.
- Patrick Adlercreutz; Immobilisation and application of lipases in organic media. Chemical Society Reviews 2013, 42, 6406-6436, 10.1039/c3cs35446f.
- Verger, R.; “Interfacial activation” of lipases: Facts and artifacts. Trends in biotechnology 1997, 15, 32-8, https://doi.org/10.1016/S0167-7799(96)10064-0.
- P. Reis; K. Holmberg; H. Watzke; M.E. Leser; R. Miller; Lipases at interfaces: A review. Advances in Colloid and Interface Science 2009, 147-148, 237-250, 10.1016/j.cis.2008.06.001.
- Robert Kourist; Henrike Brundiek; Uwe T. Bornscheuer; Protein engineering and discovery of lipases. European Journal of Lipid Science and Technology 2010, 112, 64-74, 10.1002/ejlt.200900143.
- Yuanyuan Zhang; Yuanyuan Zhao; Xin Gao; Weiwei Jiang; Zewen Li; Quancai Yao; Fengke Yang; Fanye Wang; Junhong Liu; Kinetic model of the enzymatic Michael addition for synthesis of mitomycin analogs catalyzed by immobilized lipase from T. laibacchii. Molecular Catalysis 2019, 466, 146-156, 10.1016/j.mcat.2019.01.017.
- Seizaburo Shiraga; Masaji Ishiguro; Harukazu Fukami; Masahiro Nakao; Mitsuyoshi Ueda; Creation of Rhizopus oryzae lipase having a unique oxyanion hole by combinatorial mutagenesis in the lid domain. Applied Microbiology and Biotechnology 2005, 68, 779-785, 10.1007/s00253-005-1935-0.
- Soňa Hermanová; Marie Zarevúcká; Daniel Bouša; Dr. Martin Pumera; Dr. Zdeněk Sofer; Graphene oxide immobilized enzymes show high thermal and solvent stability. Nanoscale 2015, 7, 5852-5858, 10.1039/c5nr00438a.
- Jayshree B. Kantak; Asmita Prabhune; Characterization of Smallest Active Monomeric Lipase from Novel Rhizopus Strain: Application in Transesterification. Applied Biochemistry and Biotechnology 2012, 166, 1769-1780, 10.1007/s12010-012-9584-0.
- Tigran V. Yuzbashev; Evgeniya Y. Yuzbasheva; Tatiana V. Vibornaya; Tatiana I. Sobolevskaya; Ivan A. Laptev; Alexey V. Gavrikov; Sergey P. Sineoky; Production of recombinant Rhizopus oryzae lipase by the yeast Yarrowia lipolytica results in increased enzymatic thermostability. Protein Expression and Purification 2012, 82, 83-89, 10.1016/j.pep.2011.11.014.
- H D Beer; G Wohlfahrt; R D Schmid; J E McCarthy; The folding and activity of the extracellular lipase of Rhizopus oryzae are modulated by a prosequence.. Biochemical Journal 1996, 319, 351-359.
- Afshin Ebrahimpour; Raja Rahman; Hamidon Basri; Abu Bakar Salleh; High level expression and characterization of a novel thermostable, organic solvent tolerant, 1,3-regioselective lipase from Geobacillus sp. strain ARM. Bioresource Technology 2011, 102, 6972-6981, 10.1016/j.biortech.2011.03.083.
- Shuen-Fuh Lin; Production and stabilization of a solvent-tolerant alkaline lipase from Pseudomonas pseudoalcaligenes F-111. Journal of Fermentation and Bioengineering 1996, 82, 448-451, 10.1016/s0922-338x(97)86981-4.
- Emmanuel Lesuisse; Karin Schanck; Charles Colson; Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. JBIC Journal of Biological Inorganic Chemistry 1993, 216, 155-160, 10.1111/j.1432-1033.1993.tb18127.x.
- Anjali Bose; Hareshkumar Keharia; Production, characterization and applications of organic solvent tolerant lipase by Pseudomonas aeruginosa AAU2. Biocatalysis and Agricultural Biotechnology 2013, 2, 255-266, 10.1016/j.bcab.2013.03.009.
- Jinyong Yan; Jiangke Yang; Li Xu; Yunjun Yan; Gene cloning, overexpression and characterization of a novel organic solvent tolerant and thermostable lipase from Galactomyces geotrichum Y05. Journal of Molecular Catalysis B: Enzymatic 2007, 49, 28-35, 10.1016/j.molcatb.2007.07.006.
- Heyun Zhao; Lina Zheng; Xiaofeng Wang; Yun Liu; Li Xu; Yunjun Yan; Cloning, expression and characterization of a new lipase from Yarrowia lipolytica. Biotechnology Letters 2011, 33, 2445-2452, 10.1007/s10529-011-0711-8.
- Madhu Katiyar; Amjad Ali; Effect of Metal Ions on the Hydrolytic and Transesterification Activities of Candida rugosa Lipase. Journal of Oleo Science 2013, 62, 919-924, 10.5650/jos.62.919.
- Z. Burcu Bakır Ateşlier; Kubilay Metin; Production and partial characterization of a novel thermostable esterase from a thermophilic Bacillus sp.. Enzyme and Microbial Technology 2006, 38, 628-635, 10.1016/j.enzmictec.2005.07.015.
- Wei Li; Ren-Wang Li; Qiang Li; Wei Du; Dehua Liu; Acyl migration and kinetics study of 1(3)-positional specific lipase of Rhizopus oryzae-catalyzed methanolysis of triglyceride for biodiesel production. Process Biochemistry 2010, 45, 1888-1893, 10.1016/j.procbio.2010.03.034.
- Dovilė Šinkūnienė; Patrick Adlercreutz; Effects of Regioselectivity and Lipid Class Specificity of Lipases on Transesterification, Exemplified by Biodiesel Production. Journal of the American Oil Chemists' Society 2014, 91, 1283-1290, 10.1007/s11746-014-2465-7.
- Susumu Okumura; Mieko Iwai; Yoshio Tsujisaka; Positional Specificities of Four Kinds of Microbial Lipases. Agricultural and Biological Chemistry 1976, 40, 655-660, 10.1080/00021369.1976.10862109.
- Albert Canet; Maria Dolors Benaiges; Francisco Valero; Patrick Adlercreutz; Exploring substrate specificities of a recombinant Rhizopus oryzae lipase in biodiesel synthesis. New Biotechnology 2017, 39, 59-67, 10.1016/j.nbt.2017.07.003.
- Xi Cao; Juan Mangas-Sánchez; Fengqin Feng; Patrick Adlercreutz; Acyl migration in enzymatic interesterification of triacylglycerols: Effects of lipases fromThermomyces lanuginosusandRhizopus oryzae, support material, and water activity. European Journal of Lipid Science and Technology 2016, 118, 1579-1587, 10.1002/ejlt.201500485.
- Ashok Kumar; Kartik Dhar; Shamsher Singh Kanwar; Pankaj Kumar Arora; Lipase catalysis in organic solvents: advantages and applications. Biological Procedures Online 2016, 18, 1-11, 10.1186/s12575-016-0033-2.
- A Zaks; A M Klibanov; Enzymatic catalysis in nonaqueous solvents.. Journal of Biological Chemistry 1988, 263, 3194-201.
- Mirella Di Lorenzo; Aurelio Hidalgo; Michael Haas; Ioannis V. Pavlidis Martin S. Weiß Maika Genz Uwe T. Bornscheuer; Heterologous Production of Functional Forms of Rhizopus oryzae Lipase in Escherichia coli. Applied and Environmental Microbiology 2005, 71, 8974-8977, 10.1128/aem.71.12.8974-8977.2005.
- Eda Çelik; Pınar Çalık; Production of recombinant proteins by yeast cells. Biotechnology Advances 2012, 30, 1108-1118, 10.1016/j.biotechadv.2011.09.011.
- Veeresh Juturu; Jin Chuan Wu; Heterologous Protein Expression in Pichia pastoris : Latest Research Progress and Applications. ChemBioChem 2017, 19, 7-21, 10.1002/cbic.201700460.
- Mudassar Ahmad; Melanie Hirz; Harald Pichler; Helmut Schwab; Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Applied Microbiology and Biotechnology 2014, 98, 5301-5317, 10.1007/s00253-014-5732-5.
- Xavier García-Ortega; Elena Cámara; Pau Ferrer; Joan Albiol; José Luis Montesinos-Seguí; Francisco Valero; Rational development of bioprocess engineering strategies for recombinant protein production in Pichia pastoris (Komagataella phaffii) using the methanol-free GAP promoter. Where do we stand?. New Biotechnology 2019, 53, 24-34, 10.1016/j.nbt.2019.06.002.
- Diethard Mattanovich; Alexandra B. Graf; Johannes Stadlmann; Martin Dragosits; Andreas Redl; Michael Maurer; Martin Kleinheinz; Michael Sauer; Friedrich Altmann; Brigitte Gasser; et al. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microbial Cell Factories 2009, 8, 29-29, 10.1186/1475-2859-8-29.
- Johannes Hemmerich; Núria Adelantado; José Manuel Barrigón; Xavier Ponte; Astrid Hörmann; Pau Ferrer; Frank Kensy; Francisco Valero; Comprehensive clone screening and evaluation of fed-batch strategies in a microbioreactor and lab scale stirred tank bioreactor system: application on Pichia pastoris producing Rhizopus oryzae lipase. Microbial Cell Factories 2014, 13, 36-36, 10.1186/1475-2859-13-36.
- Josu López-Fernández; Maria Dolors Benaiges; Francisco Valero; Rhizopus oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications. Catalysts 2020, 10, 1277, 10.3390/catal10111277.
- Wei Li; Wei Du; Dehua Liu; Rhizopus oryzae IFO 4697 whole cell catalyzed methanolysis of crude and acidified rapeseed oils for biodiesel production in tert-butanol system. Process Biochemistry 2007, 42, 1481-1485, 10.1016/j.procbio.2007.05.015.
- Wei Li; Wei Du; Dehua Liu; Optimization of whole cell-catalyzed methanolysis of soybean oil for biodiesel production using response surface methodology. Journal of Molecular Catalysis B: Enzymatic 2007, 45, 122-127, 10.1016/j.molcatb.2007.01.002.
- Shinji Hama; Hideki Yamaji; Masaru Kaieda; Mitsuhiro Oda; Akihiko Kondo; Hideki Fukuda; Effect of fatty acid membrane composition on whole-cell biocatalysts for biodiesel-fuel production. Biochemical Engineering Journal 2004, 21, 155-160, 10.1016/j.bej.2004.05.009.
- Matsumoto T.; Takahashi S.; Kaieda M.; Ueda M.; Tanaka A.; Fukuda H.; Kondo A.; Yeast whole-cell biocatalyst constructed by intracellular overproduction of Rhizopus oryzae lipase is applicable to biodiesel fuel production. Applied Microbiology and Biotechnology 2001, 57, 515-520, 10.1007/s002530100733.
- Takeshi Matsumoto; Shouji Takahashi; Mitsuyoshi Ueda; Atsuo Tanaka; Hideki Fukuda; Akihiko Kondo; Preparation of high activity yeast whole cell bioctalysts by optimization of intracellular production of recombinant Rhizopus oryzae lipase. Journal of Molecular Catalysis B: Enzymatic 2002, 17, 143-149, 10.1016/s1381-1177(02)00021-8.
- Takanori Tanino; Hideki Fukuda; Akihiko Kondo; Construction of a Pichia pastoris Cell-Surface Display System Using Flo1p Anchor System. Biotechnology Progress 2006, 22, 989-993, 10.1021/bp060133+.
- Wenqian Li; Hao Shi; Huaihai Ding; Liangliang Wang; Yu Zhang; Xun Li; Fei Wang; Cell Surface Display and Characterization of Rhizopus oryzae Lipase in Pichia pastoris Using Sed1p as an Anchor Protein. Current Microbiology 2015, 71, 150-155, 10.1007/s00284-015-0835-5.
- Agbo Ken Ugo; Arazu Vivian Amara; Igwe Cn; Uzo Kenechuwku; Microbial Lipases: A Prospect for Biotechnological Industrial Catalysis for Green Products: A Review. Fermentation Technology 2017, 6, 144-156, 10.4172/2167-7972.1000144.
- Stuart M Thomas; Robert DiCosimo; Vasantha Nagarajan; Biocatalysis: applications and potentials for the chemical industry. Trends in Biotechnology 2002, 20, 238-242, 10.1016/s0167-7799(02)01935-2.
- David J. Tenenbaum; Food vs. Fuel: Diversion of Crops Could Cause More Hunger. Environmental Health Perspectives 2008, 116, A254-7, 10.1289/ehp.116-a254.
- Ashraf Amin; Review of diesel production from renewable resources: Catalysis, process kinetics and technologies. Ain Shams Engineering Journal 2019, 10, 821-839, 10.1016/j.asej.2019.08.001.
- Digambar Singh; Dilip Sharma; S.L. Soni; Sumit Sharma; Pushpendra Kumar Sharma; Amit Jhalani; A review on feedstocks, production processes, and yield for different generations of biodiesel. Fuel 2020, 262, 116553, 10.1016/j.fuel.2019.116553.
- Guo Yong Yew; Sze Ying Lee; Wei-Hsin Chen; Yang Tao; Chung Lim Law; Thi Trung Chinh Nguyen; Jo-Shu Chang; Recent advances in algae biodiesel production: From upstream cultivation to downstream processing. Bioresource Technology Reports 2019, 7, 100227, 10.1016/j.biteb.2019.100227.
- Irnayuli R. Sitepu; Luis A. Garay; Ryan Sestric; David Levin; David E. Block; J. Bruce German; Kyria Boundy-Mills; Oleaginous yeasts for biodiesel: Current and future trends in biology and production. Biotechnology Advances 2014, 32, 1336-1360, 10.1016/j.biotechadv.2014.08.003.
- Digambar Singh; Dilip Sharma; S.L. Soni; Sumit Sharma; Deepika Kumari; Chemical compositions, properties, and standards for different generation biodiesels: A review. Fuel 2019, 253, 60-71, 10.1016/j.fuel.2019.04.174.
- Indu Ambat; Varsha Srivastava; Mika Sillanpää; Recent advancement in biodiesel production methodologies using various feedstock: A review. Renewable and Sustainable Energy Reviews 2018, 90, 356-369, 10.1016/j.rser.2018.03.069.
- Srivathsan Vembanur Ranganathan; Srinivasan Lakshmi Narasimhan; Karuppan Muthukumar; An overview of enzymatic production of biodiesel. Bioresource Technology 2008, 99, 3975-3981, 10.1016/j.biortech.2007.04.060.
- Abhishek Guldhe; Bhaskar Singh; Taurai Mutanda; Kugen Permaul; Faizal Bux; Advances in synthesis of biodiesel via enzyme catalysis: Novel and sustainable approaches. Renewable and Sustainable Energy Reviews 2015, 41, 1447-1464, 10.1016/j.rser.2014.09.035.
- Lew P. Christopher; Hemanathan Kumar; Vasudeo P. Zambare; Enzymatic biodiesel: Challenges and opportunities. Applied Energy 2014, 119, 497-520, 10.1016/j.apenergy.2014.01.017.
- Samuel Santos; Jaime Puna; J.F. Gomes; A Review on Bio-Based Catalysts (Immobilized Enzymes) Used for Biodiesel Production. Energies 2020, 13, 3013, 10.3390/en13113013.
- Kírian Bonet-Ragel; Albert Canet; Maria Dolors Benaiges; Francisco P J Valero; Synthesis of biodiesel from high FFA alperujo oil catalysed by immobilised lipase. Fuel 2015, 161, 12-17, 10.1016/j.fuel.2015.08.032.
- Yun-Huin Lin; Jheng-Jin Luo; Sz-Chwun John Hwang; Pei-Ru Liau; Weng-Jang Lu; Hom-Ti Lee; The influence of free fatty acid intermediate on biodiesel production from soybean oil by whole cell biocatalyst. Biomass and Bioenergy 2011, 35, 2217-2223, 10.1016/j.biombioe.2011.02.039.
- Marzieh Aghababaie; Masoud Beheshti; Amir Razmjou; Abdol-Khalegh Bordbar; Enzymatic biodiesel production from crude Eruca sativa oil using Candida rugosa lipase in a solvent-free system using response surface methodology. Biofuels 2017, 11, 93-99, 10.1080/17597269.2017.1345359.
- Abhishek Guldhe; Poonam Singh; Nirmal Renuka; Faizal Bux; Biodiesel synthesis from wastewater grown microalgal feedstock using enzymatic conversion: A greener approach. Fuel 2019, 237, 1112-1118, 10.1016/j.fuel.2018.10.033.
- Katerine S. Moreira; Lourembergue S. Moura Júnior; Rodolpho R. C. Monteiro; André L. B. De Oliveira; Camila P. Valle; Tiago M. Freire; Pierre B. A. Fechine; Maria C. M. De Souza; Gloria Fernandez-Lorente; José M. Guisán; et al.José C. S. Dos Santos Optimization of the Production of Enzymatic Biodiesel from Residual Babassu Oil (Orbignya sp.) via RSM. Catalysts 2020, 10, 414, 10.3390/catal10040414.
- Sergey N. Fedosov; Jesper Brask; Anders K. Pedersen; Mathias Nordblad; John M. Woodley; Xuebing Xu; Kinetic model of biodiesel production using immobilized lipase Candida antarctica lipase B. Journal of Molecular Catalysis B: Enzymatic 2013, 85-86, 156-168, 10.1016/j.molcatb.2012.09.011.
- Li-Hao Liu; Yung-Han Shih; Wan-Ling Liu; Chia-Her Lin; Hsi-Ya Huang; Enzyme Immobilized on Nanoporous Carbon Derived from Metal-Organic Framework: A New Support for Biodiesel Synthesis. ChemSusChem 2017, 10, 1364-1369, 10.1002/cssc.201700142.
- Shweta Shah; Munishwar N. Gupta; The effect of ultrasonic pre-treatment on the catalytic activity of lipases in aqueous and non-aqueous media. Chemistry Central Journal 2008, 2, 1-1, 10.1186/1752-153x-2-1.
- F.M. Bautista; Laura Aguado-Deblas; Felipa M. Bautista; Diego Luna; Carlos Luna; Juan Calero; Alejandro Posadillo; Antonio A. Romero; Biodiesel at the Crossroads: A Critical Review. Catalysts 2019, 9, 1033, 10.3390/catal9121033.
- Robert O. Dunn; Effects of Monoacylglycerols on the Cold Flow Properties of Biodiesel. Journal of the American Oil Chemists' Society 2012, 89, 1509-1520, 10.1007/s11746-012-2045-7.
- Juan Calero; Cristóbal Verdugo-Escamilla; Diego Luna; Enrique D. Sancho; Carlos Luna; Alejandro Posadillo; Felipa M. Bautista; Antonio A. Romero; Selective ethanolysis of sunflower oil with Lipozyme RM IM, an immobilized Rhizomucor miehei lipase, to obtain a biodiesel-like biofuel, which avoids glycerol production through the monoglyceride formation. New Biotechnology 2014, 31, 596-601, 10.1016/j.nbt.2014.02.008.
- Kírian Bonet-Ragel; Albert Canet; M Dolors Benaiges; Francisco Valero; Effect of acyl-acceptor stepwise addition strategy using alperujo oil as a substrate in enzymatic biodiesel synthesis. Journal of Chemical Technology & Biotechnology 2017, 93, 541-547, 10.1002/jctb.5399.
- Eveline Fredrick; Kim Moens; Bart Heyman; Sabine Fischer; Paul Van Der Meeren; Koen Dewettinck; Monoacylglycerols in dairy recombined cream: I. The effect on milk fat crystallization. Food Research International 2013, 51, 892-898, 10.1016/j.foodres.2013.02.007.
- Ivaldo Itabaiana; Karen M Goncalves; Y.M.L. Cordeiro; Maria Zoumpanioti; Ivana C R Leal; Leandro S M Miranda; R.O.M.A. De Souza; A. Xenakis; Kinetics and mechanism of lipase catalyzed monoacylglycerols synthesis. Journal of Molecular Catalysis B: Enzymatic 2013, 96, 34-39, 10.1016/j.molcatb.2013.06.008.
- Maria Manuela Camino Feltes; Débora De Oliveira; Jane Mara Block; Jorge Luiz Ninow; The Production, Benefits, and Applications of Monoacylglycerols and Diacylglycerols of Nutritional Interest. Food and Bioprocess Technology 2012, 6, 17-35, 10.1007/s11947-012-0836-3.
- Albert Canet; Kírian Bonet-Ragel; Maria Dolors Benaiges; Francisco Valero; Biodiesel synthesis in a solvent-free system by recombinant Rhizopus oryzae: comparative study between a stirred tank and a packed-bed batch reactor. Biocatalysis and Biotransformation 2017, 35, 35-40, 10.1080/10242422.2016.1278211.
- Gustavo Ciudad; Isaac Reyes; Milko A Jorquera; Laura Azócar; Lukas Y. Wick; Rodrigo Navia; Novel three-phase bioreactor concept for fatty acid alkyl ester production using R. oryzae as whole cell catalyst. World Journal of Microbiology and Biotechnology 2011, 27, 2505-2512, 10.1007/s11274-011-0719-1.
- Myung Gwi Jang; Deog Keun Kim; Soon Chul Park; Jin Suk Lee; Seung Wook Kim; Biodiesel production from crude canola oil by two-step enzymatic processes. Renewable Energy 2012, 42, 99-104, 10.1016/j.renene.2011.09.009.
- Qiyang He; Hao Shi; Huaxiang Gu; Gilda Naka; Huaihai Ding; Xun Li; Yu Zhang; Bo Hu; Fei Wang; Immobilization of Rhizopus oryzae LY6 onto Loofah Sponge as a Whole-Cell Biocatalyst for Biodiesel Production. BioResources 2015, 11, 850-860, 10.15376/biores.11.1.850-860.
- Feng Su; Guan-Lin Li; Yan-Li Fan; Yun-Jun Yan; Enhancing biodiesel production via a synergic effect between immobilized Rhizopus oryzae lipase and Novozym 435. Fuel Processing Technology 2015, 137, 298-304, 10.1016/j.fuproc.2015.03.013.
- Gui-Xiong Zhou; Guanyi Chen; Bei-Bei Yan; Biodiesel production in a magnetically-stabilized, fluidized bed reactor with an immobilized lipase in magnetic chitosan microspheres. Biotechnology Letters 2013, 36, 63-68, 10.1007/s10529-013-1336-x.
- Shinji Hama; Hideki Yamaji; Takahiro Fukumizu; Takao Numata; Sriappareddy Tamalampudi; Akihiko Kondo; Hideo Noda; Hideki Fukuda; Biodiesel-fuel production in a packed-bed reactor using lipase-producing Rhizopus oryzae cells immobilized within biomass support particles. Biochemical Engineering Journal 2007, 34, 273-278, 10.1016/j.bej.2006.12.013.
- Carlos Luna; Cristóbal Verdugo-Escamilla; Enrique D. Sancho; Diego Luna; Juan Calero; Alejandro Posadillo; Felipa M. Bautista; Antonio A. Romero; Biocatalytic Behaviour of Immobilized Rhizopus oryzae Lipase in the 1,3-Selective Ethanolysis of Sunflower Oil to Obtain a Biofuel Similar to Biodiesel. Molecules 2014, 19, 11419-11439, 10.3390/molecules190811419.
- Carlos Luna; Cristóbal Verdugo-Escamilla; Enrique D. Sancho; Diego Luna; Juan Calero; Alejandro Posadillo; Felipa M. Bautista; Antonio A. Romero; A Biofuel Similar to Biodiesel Obtained by Using a Lipase from Rhizopus oryzae, Optimized by Response Surface Methodology. Energies 2014, 7, 3383-3399, 10.3390/en7053383.
- L.C. Meher; C.P. Churamani; Arif; Zakwan Ahmed; S.N. Naik; Jatropha curcas as a renewable source for bio-fuels—A review. Renewable and Sustainable Energy Reviews 2013, 26, 397-407, 10.1016/j.rser.2013.05.065.
- Joana Rodrigues; Véronique Perrier; Jérôme LeComte; Eric Dubreucq; Suzana Ferreira-Dias; Biodiesel production from crude jatropha oil catalyzed by immobilized lipase/acyltransferase from Candida parapsilosis in aqueous medium. Bioresource Technology 2016, 218, 1224-1229, 10.1016/j.biortech.2016.07.090.
- Xun Li; Xiao-Yun He; Zhi-Lin Li; You-Dong Wang; Chun-Yu Wang; Hao Shi; Fei Wang; Enzymatic production of biodiesel from Pistacia chinensis bge seed oil using immobilized lipase. Fuel 2012, 92, 89-93, 10.1016/j.fuel.2011.06.048.
- A. Arumugam; V. Ponnusami; Biodiesel production from Calophyllum inophyllum oil using lipase producing Rhizopus oryzae cells immobilized within reticulated foams. Renewable Energy 2014, 64, 276-282, 10.1016/j.renene.2013.11.016.
- Elvira Navarro López; Alfonso Robles Medina; Pedro Antonio González Moreno; Luis Esteban Cerdán; Lorena Martín Valverde; Emilio Molina Grima; Biodiesel production from Nannochloropsis gaditana lipids through transesterification catalyzed by Rhizopus oryzae lipase. Bioresource Technology 2016, 203, 236-244, 10.1016/j.biortech.2015.12.036.
- Elvira Navarro López; Alfonso Robles; Pedro Antonio González Moreno; Luis Esteban Cerdán; E. Molina-Grima; Extraction of microalgal lipids and the influence of polar lipids on biodiesel production by lipase-catalyzed transesterification. Bioresource Technology 2016, 216, 904-913, 10.1016/j.biortech.2016.06.035.
- K. Araya; A. Ugarte; Laura Azocar; O. Valerio; Lukas Y Wick; Gustavo Ciudad; Whole cell three phase bioreactors allow for effective production of fatty acid alkyl esters derived from microalgae lipids. Fuel 2015, 144, 25-32, 10.1016/j.fuel.2014.12.014.
- Tahereh Nematian; Zeinab Salehi; Alireza Shakeri; Conversion of bio-oil extracted from Chlorella vulgaris micro algae to biodiesel via modified superparamagnetic nano-biocatalyst. Renewable Energy 2020, 146, 1796-1804, 10.1016/j.renene.2019.08.048.
- Papasanee Muanruksa; Pakawadee Kaewkannetra; Combination of fatty acids extraction and enzymatic esterification for biodiesel production using sludge palm oil as a low-cost substrate. Renewable Energy 2020, 146, 901-906, 10.1016/j.renene.2019.07.027.
- Sanjib Kumar Karmee; Wian Swanepoel; Sanette Marx; Biofuel production from spent coffee grounds via lipase catalysis. Energy Sources, Part A: Recovery, Utilization, and Environmental Effects 2017, 40, 294-300, 10.1080/15567036.2017.1415394.
- Bharathiraja, B.; Ranjithkumar, R.; Chakravarthy, M.; Yogendran, D.; Vivek, P.; Yuvaraj, D.; Kumar, R.P.; Palani, S.; Kinetic analysis of fatty acid alkyl esters using whole cell biocatalyst and lipase catalyzed transesterification from waste cooking oil. Asian J. Microbiol. Biotechnol. Environ. Sci. 2014, 16, 745-752.
- Mahin Basha Syed; Mohammed Yousuf Ali; Mohammed Ishaq; S. Bakkiyaraj; M.G. Devanesan; Viruthagiri Tangavelu; Response surface optimization of biodiesel production using immobilized Rhizopus oryzae cells. Biofuels 2016, 7, 457-464, 10.1080/17597269.2016.1153364.
- Ting Sun; Wei Du; Dehua Liu; Comparative study on stability of whole cells during biodiesel production in solvent-free system. Process Biochemistry 2011, 46, 661-664, 10.1016/j.procbio.2010.11.006.
- Kazuhiro Ban; Shinji Hama; Keiko Nishizuka; Masaru Kaieda; Takeshi Matsumoto; Akihiko Kondo; Hideo Noda; Hideki Fukuda; Repeated use of whole-cell biocatalysts immobilized within biomass support particles for biodiesel fuel production. Journal of Molecular Catalysis B: Enzymatic 2002, 17, 157-165, 10.1016/s1381-1177(02)00023-1.
- Kírian Bonet-Ragel; Lucia López-Pou; Gisela Tutusaus; Maria Dolors Benaiges; Francisco Valero; Rice husk ash as a potential carrier for the immobilization of lipases applied in the enzymatic production of biodiesel. Biocatalysis and Biotransformation 2017, 36, 151-158, 10.1080/10242422.2017.1308498.
- Susan Hartwig Duarte; Gonzalo Lázaro Del Peso Hernández; Albert Canet; Maria Dolors Benaiges; Francisco Maugeri; Francisco Valero; Enzymatic biodiesel synthesis from yeast oil using immobilized recombinant Rhizopus oryzae lipase. Bioresource Technology 2015, 183, 175-180, 10.1016/j.biortech.2015.01.133.
- Feng Su; Guanlin Li; Houjin Zhang; Yunjun Yan; Enhanced Performance of Rhizopus oryzae Lipase Immobilized on Hydrophobic Carriers and Its Application in Biorefinery of Rapeseed Oil Deodorizer Distillate. BioEnergy Research 2014, 7, 935-945, 10.1007/s12155-014-9415-y.
- S. Bakkiyaraj; Mahin Basha Syed; M. G. Devanesan; Viruthagiri Thangavelu; Production and optimization of biodiesel using mixed immobilized biocatalysts in packed bed reactor. Environmental Science and Pollution Research 2015, 23, 9276-9283, 10.1007/s11356-015-4583-7.
- Bharathiraja Balasubramanian; Praveen Kumar Ramanujam; Ranjith Ravi Kumar; Chakravarthy Muninathan; Yogendran Dhinakaran; Optimization of biological transesterification of waste cooking oil in different solvents using response surface methodology. Management of Environmental Quality: An International Journal 2016, 27, 537-550, 10.1108/meq-06-2015-0118.
- Tahereh Nematian; Alireza Shakeri; Zeinab Salehi; Ali Akbar Saboury; Lipase immobilized on functionalized superparamagnetic few-layer graphene oxide as an efficient nanobiocatalyst for biodiesel production from Chlorella vulgaris bio-oil. Biotechnology for Biofuels 2020, 13, 57, 10.1186/s13068-020-01688-x.
- Sneha Athalye; Ratna R Sharmashivappa; Steven W Peretti; Praveen Kolar; Jack P. Davis; Producing biodiesel from cottonseed oil using Rhizopus oryzae ATCC #34612 whole cell biocatalysts: Culture media and cultivation period optimization. Energy for Sustainable Development 2013, 17, 331-336, 10.1016/j.esd.2013.03.009.
- V.C. Vipin; Jilse Sebastian; C. Muraleedharan; A. Santhiagu; Enzymatic Transesterification of Rubber Seed Oil Using Rhizopus Oryzae Lipase. Procedia Technology 2016, 25, 1014-1021, 10.1016/j.protcy.2016.08.201.
- Jong Ho Lee; Sung Bong Kim; Seong Woo Kang; Yoon Seok Song; Chulhwan Park; Sung Ok Han; Seung Wook Kim; Biodiesel production by a mixture of Candida rugosa and Rhizopus oryzae lipases using a supercritical carbon dioxide process. Bioresource Technology 2011, 102, 2105-2108, 10.1016/j.biortech.2010.08.034.
- Leping Zeng; Yaojia He; Liangcheng Jiao; Kai Li; Yunjun Yan; Preparation of Biodiesel with Liquid Synergetic Lipases from Rapeseed Oil Deodorizer Distillate. Applied Biochemistry and Biotechnology 2017, 183, 778-791, 10.1007/s12010-017-2463-y.
- J. Rodrigues; A. Canet; I. Rivera; N.M. Osório; G. Sandoval; F. Valero; Suzana Ferreira-Dias; Biodiesel production from crude Jatropha oil catalyzed by non-commercial immobilized heterologous Rhizopus oryzae and Carica papaya lipases. Bioresource Technology 2016, 213, 88-95, 10.1016/j.biortech.2016.03.011.
- Hamid Mukhtar; Samreen Khursheed; Ikram- Ul- Haq; Muhammad Waseem Mumtaz; Umer Rashid; Saud Ibrahim Al-Resayes; Optimization of Lipase Biosynthesis fromRhizopus oryzaefor Biodiesel Production Using Multiple Oils. Chemical Engineering & Technology 2016, 39, 1707-1715, 10.1002/ceat.201500584.
- Gui-Xiong Zhou; Guanyi Chen; Bei-Bei Yan; Two-step biocatalytic process using lipase and whole cell catalysts for biodiesel production from unrefined jatropha oil. Biotechnology Letters 2015, 37, 1959-1963, 10.1007/s10529-015-1883-4.
- S. Sattari; F. Vahabzadeh; H. K. Aghtaei; PERFORMANCE OF LOOFA-IMMOBILIZED Rhizopus oryzae IN THE ENZYMATIC PRODUCTION OF BIODIESEL WITH USE OF OLEIC ACID IN n-HEXANE MEDIUM. Brazilian Journal of Chemical Engineering 2015, 32, 367-376, 10.1590/0104-6632.20150322s00003525.
- Prashanth Ramachandran; Guru Krupa Narayanan; Sakthivel Gandhi; Swaminathan Sethuraman; Uma Maheswari Krishnan; Rhizopus oryzae Lipase Immobilized on Hierarchical Mesoporous Silica Supports for Transesterification of Rice Bran Oil. Applied Biochemistry and Biotechnology 2014, 175, 2332-2346, 10.1007/s12010-014-1432-y.
- Alireza Zarei; Nor Aishah Saidina Amin; Amin Talebian-Kiakalaieh; Nor Azimah Mohd Zain; Immobilized lipase-catalyzed transesterification of Jatropha curcas oil: Optimization and modeling. Journal of the Taiwan Institute of Chemical Engineers 2014, 45, 444-451, 10.1016/j.jtice.2013.05.015.
- Albert Canet; Maria Dolors Benaiges; Francisco Valero; Biodiesel Synthesis in a Solvent-Free System by Recombinant Rhizopus oryzae Lipase. Study of the Catalytic Reaction Progress. Journal of the American Oil Chemists' Society 2014, 91, 1499-1506, 10.1007/s11746-014-2498-y.
- Grazielle S. S. Andrade; Larissa Freitas; Pedro C. Oliveira; Heizir F. De Castro; Screening, immobilization and utilization of whole cell biocatalysts to mediate the ethanolysis of babassu oil. Journal of Molecular Catalysis B: Enzymatic 2012, 84, 183-188, 10.1016/j.molcatb.2012.02.011.
- Suchit Deshmukh; Ritunesh Kumar; Kiran Bala; Microalgae biodiesel: A review on oil extraction, fatty acid composition, properties and effect on engine performance and emissions. Fuel Processing Technology 2019, 191, 232-247, 10.1016/j.fuproc.2019.03.013.
- Zhihong Yin; Liandong Zhu; ShuangXi Li; Tianyi Hu; Ruoyu Chu; Fan Mo; Dan Hu; Chenchen Liu; Bin Li; A comprehensive review on cultivation and harvesting of microalgae for biodiesel production: Environmental pollution control and future directions. Bioresource Technology 2020, 301, 122804, 10.1016/j.biortech.2020.122804.
- Pedro F Lisboa; Ana Rita Rodrigues; José Luis Martín; Pedro C Simoes; Susana Barreiros; Alexandre Paiva; Economic analysis of a plant for biodiesel production from waste cooking oil via enzymatic transesterification using supercritical carbon dioxide. The Journal of Supercritical Fluids 2014, 85, 31-40, 10.1016/j.supflu.2013.10.018.
- Theocharis Tsoutsos; Stavroula Tournaki; Zacharias Gkouskos; Orlando Paraíba; Filippo Giglio; Pablo Quero García; João Braga; Haris Adrianos; Monica Filice; Quality Characteristics of Biodiesel Produced from Used Cooking Oil in Southern Europe. ChemEngineering 2019, 3, 19, 10.3390/chemengineering3010019.
- Zahira Yaakob; Masita Mohammad; Mohammad Alherbawi; Zahangir Alam; Kamaruzaman Sopian; Overview of the production of biodiesel from Waste cooking oil. Renewable and Sustainable Energy Reviews 2013, 18, 184-193, 10.1016/j.rser.2012.10.016.
- Carlos Daniel Mandolesi De Araújo; Claudia Cristina De Andrade; Erika De Souza E Silva; Francisco Antonio Dupas; Biodiesel production from used cooking oil: A review. Renewable and Sustainable Energy Reviews 2013, 27, 445-452, 10.1016/j.rser.2013.06.014.
- Y Zhang; M.A Dubé; D.D McLean; M Kates; Biodiesel production from waste cooking oil: 2. Economic assessment and sensitivity analysis. Bioresource Technology 2003, 90, 229-240, 10.1016/s0960-8524(03)00150-0.
- Alessandra Basso; Simona Serban; Industrial applications of immobilized enzymes—A review. Molecular Catalysis 2019, 479, 110607, 10.1016/j.mcat.2019.110607.
- Roswanira Abdul Wahab; Nursyafiqah Elias; Faizuan Abdullah; Sib Krishna Ghoshal; On the taught new tricks of enzymes immobilization: An all-inclusive overview. Reactive and Functional Polymers 2020, 152, 104613, 10.1016/j.reactfunctpolym.2020.104613.
- Roger A. Sheldon; John M. Woodley; Role of Biocatalysis in Sustainable Chemistry. Chemical Reviews 2017, 118, 801-838, 10.1021/acs.chemrev.7b00203.
- H. Fukuda; S. Hama; S. Tamalampudi; H. Noda; Whole-cell biocatalysts for biodiesel fuel production. Trends in Biotechnology 2008, 26, 668-673, 10.1016/j.tibtech.2008.08.001.
- Roberto Fernandez-Lafuente; Pilar Armisén; Pilar Sabuquillo; Gloria Fernández-Lorente; Jose M Guisan; Immobilization of lipases by selective adsorption on hydrophobic supports. Chemistry and Physics of Lipids 1998, 93, 185-197, 10.1016/s0009-3084(98)00042-5.
- Gloria Fernandez-Lorente; Javier Rocha-Martín; Jose M Guisan; Immobilization of Lipases by Adsorption on Hydrophobic Supports: Modulation of Enzyme Properties in Biotransformations in Anhydrous Media. Methods in Molecular Biology 2020, 2100, 143-158, 10.1007/978-1-0716-0215-7_9.
- You-Dong Wang; Xiao-Yong Shen; Zhi-Lin Li; Xun Li; Fei Wang; Xiao-An Nie; Jian-Chun Jiang; Immobilized recombinant Rhizopus oryzae lipase for the production of biodiesel in solvent free system. Journal of Molecular Catalysis B: Enzymatic 2010, 67, 45-51, 10.1016/j.molcatb.2010.07.004.
- Tianwei Tan; Jike Lu; Kaili Nie; Li Deng; Fang Wang; Biodiesel production with immobilized lipase: A review. Biotechnology Advances 2010, 28, 628-634, 10.1016/j.biotechadv.2010.05.012.
- Balasubramaniyan Bharathiraja; Ayyappasamy Sudalaiyadum Perumal; Jayamuthunagai Jayaraman; Jayakumar Mani; Praveenkumar Ramanujam; Comparative analysis for the production of fatty acid alkyl esterase using whole cell biocatalyst and purified enzyme from Rhizopus oryzae on waste cooking oil (sunflower oil). Waste Management 2012, 32, 1539-1547, 10.1016/j.wasman.2012.03.011.
- Xuebing Zhao; Feng Qi; Chongli Yuan; Wei Du; Dehua Liu; Lipase-catalyzed process for biodiesel production: Enzyme immobilization, process simulation and optimization. Renewable and Sustainable Energy Reviews 2015, 44, 182-197, 10.1016/j.rser.2014.12.021.
- Adriano A. Mendes; Larissa Freitas; Ana Karine F. De Carvalho; Pedro C. De Oliveira; Heizir F. De Castro; Immobilization of a Commercial Lipase fromPenicillium camembertii(Lipase G) by Different Strategies. Enzyme Research 2011, 2011, 1-8, 10.4061/2011/967239.
- B. Norjannah; Hwai Chyuan Ong; H. H. Masjuki; J. C. Juan; W. T. Chong; Enzymatic transesterification for biodiesel production: a comprehensive review. RSC Advances 2016, 6, 60034-60055, 10.1039/c6ra08062f.
- Marina Lotti; Jürgen Pleiss; Francisco Valero; Pau Ferrer; Effects of methanol on lipases: Molecular, kinetic and process issues in the production of biodiesel. Biotechnology Journal 2014, 10, 22-30, 10.1002/biot.201400158.
- Francisco Valero; Suzana Ferreira-Dias; Georgina Sandoval; Francisco J. Plou; Maria Suzana Leitão Ferreira Dias Vicente; The potential use of lipases in the production of fatty acid derivatives for the food and nutraceutical industries. Electronic Journal of Biotechnology 2013, 16, 0, 10.2225/vol16-issue3-fulltext-5.
- Yalong Guo; Zhixiang Cai; Yanping Xie; Aiqin Ma; HongBin Zhang; Pingfan Rao; Qiang Wang; Synthesis, physicochemical properties, and health aspects of structured lipids: A review. Comprehensive Reviews in Food Science and Food Safety 2020, 19, 759-800, 10.1111/1541-4337.12537.
- Robert E. Smith; John W. Finley; Gilbert A. Leveille; Overview of SALATRIM: A family of low-calorie fats. Journal of Agricultural and Food Chemistry 1994, 42, 432-434, 10.1021/jf00038a036.
- H.T. Osborn; C.C. Akoh; Structured Lipids-Novel Fats with Medical, Nutraceutical, and Food Applications. Comprehensive Reviews in Food Science and Food Safety 2006, 1, 110-120, 10.1111/j.1541-4337.2002.tb00010.x.
- A López-López; A.I. Castellote; C Campoy-Folgoso; M Rivero-Urgël; R Tormo-Carnicé; D Infante-Pina; M.C López-Sabater; The influence of dietary palmitic acid triacylglyceride position on the fatty acid, calcium and magnesium contents of at term newborn faeces. Early Human Development 2001, 65, S83-S94, 10.1016/s0378-3782(01)00210-9.
- Neşe Şahín; Casimir C. Akoh; Artemi̇s Karaalí; Human Milk Fat Substitutes Containing Omega-3 Fatty Acids. Journal of Agricultural and Food Chemistry 2006, 54, 3717-3722, 10.1021/jf053103f.
- Nirupam Biswas; Yuen Lin Cheow; Chin Ping Tan; Lee Fong Siow; Physicochemical Properties of Enzymatically Produced Palm-Oil-Based Cocoa Butter Substitute (CBS) With Cocoa Butter Mixture. European Journal of Lipid Science and Technology 2018, 120, 1700205, 10.1002/ejlt.201700205.
- Juste Yamoneka; Paul Malumba; Georges Lognay; François Béra; Christophe Blecker; Sabine Danthine; Enzymatic Inter-Esterification of Binary Blends ContainingIrvingia gabonensisSeed Fat to Produce Cocoa Butter Substitute. European Journal of Lipid Science and Technology 2018, 120, 1700423, 10.1002/ejlt.201700423.
- Ruttiya Lakum; Sopark Sonwai; Production of trans-free margarine fat by enzymatic interesterification of soy bean oil, palm stearin and coconut stearin blend. International Journal of Food Science & Technology 2018, 53, 2761-2769, 10.1111/ijfs.13888.
- Ying Li; Jinli Zhao; Xiaodong Xie; Zhen Zhang; Ning Zhang; Yong Wang; A low trans margarine fat analog to beef tallow for healthier formulations: Optimization of enzymatic interesterification using soybean oil and fully hydrogenated palm oil. Food Chemistry 2018, 255, 405-413, 10.1016/j.foodchem.2018.02.086.
- Akiko Kawashima; Yuji Shimada; Miwa Yamamoto; Akio Sugihara; Toshihiro Nagao; Sadao Komemushi; Yoshio Tominaga; Enzymatic synthesis of high-purity structured lipids with caprylic acid at 1,3-positions and polyunsaturated fatty acid at 2-position. Journal of the American Oil Chemists' Society 2001, 78, 611-616, 10.1007/s11746-001-0313-0.
- Seong-Koon Lo; Chin-Ping Tan; Kamariah Long; Mohd. Suria Affandi Yusoff; Oi-Ming Lai; Diacylglycerol Oil—Properties, Processes and Products: A Review. Food and Bioprocess Technology 2008, 1, 223-233, 10.1007/s11947-007-0049-3.
- Lydia Fomuso; Subramani Sellappan; Casimir Akoh; Vivienne Yankah; Enzymatic Synthesis of Structured Lipids. Lipid Biotechnology 2002, 10, 129-140, 10.1201/9780203908198.ch21.
- Byung Hee Kim; Casimir C. Akoh; Recent Research Trends on the Enzymatic Synthesis of Structured Lipids. Journal of Food Science 2015, 80, C1713-C1724, 10.1111/1750-3841.12953.
- Qabul Dinanta Utama; Azis Boing Sitanggang; Dede Robiatul Adawiyah; Purwiyatno Hariyadi; Lipase-Catalyzed Interesterification for the Synthesis of Medium-Long-Medium (MLM) Structured Lipids. Food Technology and Biotechnology 2019, 57, 305-318, 10.17113/ftb.57.03.19.6025.
- Patrícia A. Nunes; Paula Pires Cabral; M. Guillén; Fernando Parrilla Valero; D. Luna; Suzana Ferreira-Dias; Production of MLM-Type Structured Lipids Catalyzed by Immobilized Heterologous Rhizopus oryzae Lipase. Journal of the American Oil Chemists' Society 2010, 88, 473-480, 10.1007/s11746-010-1702-y.
- Danyelle A. Mota; Devi Rajan; Giuditta C. Heinzl; Natália M. Osório; Jorge Gominho; Laiza C. Krause; Cleide M.F. Soares; K. Madhavan Nampoothiri; Rajeev K. Sukumaran; Suzana Ferreira-Dias; et al. Production of low-calorie structured lipids from spent coffee grounds or olive pomace crude oils catalyzed by immobilized lipase in magnetic nanoparticles. Bioresource Technology 2020, 307, 123223, 10.1016/j.biortech.2020.123223.
- Carolina M. Costa; Natália M. Osório; Albert Canet; Ivanna Rivera; Georgina Sandoval; Francisco Valero; Suzana Ferreira-Dias; Production of MLM Type Structured Lipids From Grapeseed Oil Catalyzed by Non-Commercial Lipases. European Journal of Lipid Science and Technology 2017, 120, 1700320–1700328, 10.1002/ejlt.201700320.
- Toshihiro Nagao; Akiko Kawashima; Motoo Sumida; Yomi Watanabe; Kengo Akimoto; Harukazu Fukami; Akio Sugihara; Yuji Shimada; Production of structured TAG rich in 1,3-capryloyl-2-arachidonoyl glycerol fromMortierellasingle-cell oil. Journal of the American Oil Chemists' Society 2003, 80, 867-872, 10.1007/s11746-003-0787-9.
- Patrícia A. Nunes; Paula Pires Cabral; Magno R Guillen; Francisco Valero; Suzana Ferreiradias; Batch operational stability of immobilized heterologous Rhizopus oryzae lipase during acidolysis of virgin olive oil with medium-chain fatty acids. Biochemical Engineering Journal 2012, 67, 265-268, 10.1016/j.bej.2012.06.004.
- Patrícia A. Nunes; Paula Pires Cabral; M. Guillén; Francisco Valero; Suzana Ferreiradias; Optimized Production of MLM Triacylglycerols Catalyzed by Immobilized Heterologous Rhizopus oryzae Lipase. Journal of the American Oil Chemists' Society 2012, 89, 1287-1295, 10.1007/s11746-012-2027-9.
- 267. Balieiro, A.L.; Osório, N.M.; Lima, Á.S.; Soares, C.M.F.; Valero, F.; Ferreira-Dias, S; Production of dietetic triacylglycerols from olive oil catalyzed by immobilized heterologous Rhizopus oryzae lipase. Chem. Eng. Trans 2018, 64, 0.
- Luis Esteban; María J. Jiménez; Estrella Hita; Pedro A. González; Lorena Martín; Alfonso Robles Medina; Production of structured triacylglycerols rich in palmitic acid at sn-2 position and oleic acid at sn-1,3 positions as human milk fat substitutes by enzymatic acidolysis. Biochemical Engineering Journal 2011, 54, 62-69, 10.1016/j.bej.2011.01.009.
- Tiago Simões; Francisco Valero; Carla Tecelão; Suzana Ferreira-Dias; Production of Human Milk Fat Substitutes Catalyzed by a Heterologous Rhizopus oryzae Lipase and Commercial Lipases. Journal of the American Oil Chemists' Society 2013, 91, 411-419, 10.1007/s11746-013-2379-9.
- Ana Rita Faustino; Natália M. Osório; Carla Tecelão; Albert Canet; Francisco Valero; Suzana Ferreira-Dias; Camelina oil as a source of polyunsaturated fatty acids for the production of human milk fat substitutes catalyzed by a heterologousRhizopus oryzaelipase. European Journal of Lipid Science and Technology 2015, 118, 532-544, 10.1002/ejlt.201500003.
- María Del Mar Muñío; Alfonso Robles; Luis Esteban; Pedro A. González; Emilio Molina; Synthesis of structured lipids by two enzymatic steps: Ethanolysis of fish oils and esterification of 2-monoacylglycerols. Process Biochemistry 2009, 44, 723-730, 10.1016/j.procbio.2009.03.002.
- Estrella Hita; Alfonso Robles Medina; Belén Camacho; Antonio Ramírez; Luis Esteban; María J. Jiménez; María M. Muñío; Pedro A. González; Emilio Molina; Production of structured triacylglycerols (STAG) rich in docosahexaenoic acid (DHA) in position 2 by acidolysis of tuna oil catalyzed by lipases. Process Biochemistry 2007, 42, 415-422, 10.1016/j.procbio.2006.09.023.
- Alicia Rodríguez; Luis Esteban; Lorena Martín; María José Jiménez; Estrella Hita; Beatriz Castillo; Pedro A. González; Alfonso Robles; Synthesis of 2-monoacylglycerols and structured triacylglycerols rich in polyunsaturated fatty acids by enzyme catalyzed reactions. Enzyme and Microbial Technology 2012, 51, 148-155, 10.1016/j.enzmictec.2012.05.006.
- Dequan Zhou; Xuebing Xu; Huiling Mu; Carl-Erik Høy; Jens Adler-Nissen; LIPASE-CATALYZED PRODUCTION OF STRUCTURED LIPIDS VIA ACIDOLYSIS OF FISH OIL WITH CAPRYLIC ACID. Journal of Food Lipids 2007, 7, 263-274, 10.1111/j.1745-4522.2000.tb00177.x.
- Ariela V. Paula; Gisele F. M. Nunes; Heizir F. De Castro; J.C. Dos Santos; Synthesis of Structured Lipids by Enzymatic Interesterification of Milkfat and Soybean Oil in a Basket-Type Stirred Tank Reactor. Industrial & Engineering Chemistry Research 2015, 54, 1731-1737, 10.1021/ie503189e.
- Joydeep Ray; Zoltan K. Nagy; Kevin W. Smith; Krishnadath Bhaggan; Andrew G.F. Stapley; Kinetic study of the acidolysis of high oleic sunflower oil with stearic–palmitic acid mixtures catalysed by immobilised Rhizopus oryzae lipase. Biochemical Engineering Journal 2013, 73, 17-28, 10.1016/j.bej.2012.12.018.
- T Yang; M Fruekilde; X Xu; Suppression of acyl migration in enzymatic production of structured lipids through temperature programming. Food Chemistry 2005, 92, 101-107, 10.1016/j.foodchem.2004.07.007.
- Amanda Gomes Almeida Sá; Alessandra Cristina De Meneses; Pedro Henrique Hermes De Araújo; Débora De Oliveira; A review on enzymatic synthesis of aromatic esters used as flavor ingredients for food, cosmetics and pharmaceuticals industries. Trends in Food Science & Technology 2017, 69, 95-105, 10.1016/j.tifs.2017.09.004.
- Wenyuan Gao; Kai Wu; Lifeng Chen; Haiyang Fan; Zhiqiang Zhao; Bei Gao; Hualei Wang; Dongzhi Wei; A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microbial Cell Factories 2016, 15, 41, 10.1186/s12934-016-0435-5.
- Shang-Ming Huang; Hsin-Yi Huang; Yu-Min Chen; Chia-Hung Kuo; Chwen-Jen Shieh; Continuous Production of 2-Phenylethyl Acetate in a Solvent-Free System Using a Packed-Bed Reactor with Novozym® 435. Catalysts 2020, 10, 714, 10.3390/catal10060714.
- Ikram Bayout; Nassima Bouzemi; Na Guo; Xiangzhao Mao; Stefano Serra; Sergio Riva; Francesco Secundo; Natural flavor ester synthesis catalyzed by lipases. Flavour and Fragrance Journal 2019, 35, 209-218, 10.1002/ffj.3554.
- Shamoon Asmat; Qayyum Husain; A robust nanobiocatalyst based on high performance lipase immobilized to novel synthesised poly(o-toluidine) functionalized magnetic nanocomposite: Sterling stability and application. Materials Science and Engineering: C 2019, 99, 25-36, 10.1016/j.msec.2019.01.070.
- Shamoon Asmat; Abdul Hakeem Anwer; Qayyum Husain; Immobilization of lipase onto novel constructed polydopamine grafted multiwalled carbon nanotube impregnated with magnetic cobalt and its application in synthesis of fruit flavours. International Journal of Biological Macromolecules 2019, 140, 484-495, 10.1016/j.ijbiomac.2019.08.086.
- Wellington Correa Moreira; Alfredo Luís Pereira Elias; Wislei Riuper Osório; Giovana Silva Padilha; Alternative method to improve the ethyl valerate yield using an immobilised Burkholderia cepacia lipase. Journal of Microencapsulation 2019, 36, 327-337, 10.1080/02652048.2019.1626927.
- José M. Rodriguez-Nogales; Elena Roura; Elizabeth Contreras; Biosynthesis of ethyl butyrate using immobilized lipase: a statistical approach. Process Biochemistry 2005, 40, 63-68, 10.1016/j.procbio.2003.11.049.
- Marina Guillén; Maria Dolors Benaiges; Francisco Valero; Biosynthesis of ethyl butyrate by immobilized recombinant Rhizopus oryzae lipase expressed in Pichia pastoris. Biochemical Engineering Journal 2012, 65, 1-9, 10.1016/j.bej.2012.03.009.
- Marina Guillén; Maria Dolors Benaiges; Francisco Valero; Improved ethyl butyrate synthesis catalyzed by an immobilized recombinant Rhizopus oryzae lipase: A comprehensive statistical study by production, reaction rate and yield analysis. Journal of Molecular Catalysis B: Enzymatic 2016, 133, S371-S376, 10.1016/j.molcatb.2017.02.010.
- C. Grosso; S. Ferreira-Dias; P. Pires-Cabral; Modelling and optimization of ethyl butyrate production catalysed by Rhizopus oryzae lipase. Journal of Food Engineering 2013, 115, 475-480, 10.1016/j.jfoodeng.2012.08.001.
- Riadh Ben Salah; Hanen Ghamghui; Nabil Miled; Hafedh Mejdoub; Youssef Gargouri; Production of butyl acetate ester by lipase from novel strain of Rhizopus oryzae. Journal of Bioscience and Bioengineering 2007, 103, 368-372, 10.1263/jbb.103.368.
- 290. Kumari, A.; Mahapatra, P.; Garlapati, V.K.; Banerjee, R.; Dasgupta, S; Lipase mediated isoamyl acetate synthesis in solvent-free system using vinyl acetate as acyl donor. Food Technol. Biotechnol 2009, 47, 13-18.
- Hanen Ghamgui; Maha Karra-Chaâbouni; Sofiane Bezzine; Nabil Miled; Youssef Gargouri; Production of isoamyl acetate with immobilized Staphylococcus simulans lipase in a solvent-free system. Enzyme and Microbial Technology 2006, 38, 788-794, 10.1016/j.enzmictec.2005.08.011.
- Vijay Kumar Garlapati; Rintu Banerjee; Solvent-Free Synthesis of Flavour Esters through Immobilized Lipase Mediated Transesterification. Enzyme Research 2013, 2013, 1-6, 10.1155/2013/367410.
- Kishor P. Dhake; Krishna M. Deshmukh; Yogesh P. Patil; Rekha S. Singhal; Bhalchandra M. Bhanage; Improved activity and stability of Rhizopus oryzae lipase via immobilization for citronellol ester synthesis in supercritical carbon dioxide. Journal of Biotechnology 2011, 156, 46-51, 10.1016/j.jbiotec.2011.08.019.
- Nathalia Saraiva Rios; Bruna Bandeira Pinheiro; Maísa Pessoa Pinheiro; Rayanne Mendes Bezerra; José Cleiton Sousa Dos Santos; Luciana Rocha Barros Gonçalves; Biotechnological potential of lipases from Pseudomonas: Sources, properties and applications. Process Biochemistry 2018, 75, 99-120, 10.1016/j.procbio.2018.09.003.
- Gerald Kirchner; Mark P. Scollar; Alexander M. Klibanov; Resolution of racemic mixtures via lipase catalysis in organic solvents. Journal of the American Chemical Society 1985, 107, 7072-7076, 10.1021/ja00310a052.
- Jose M. Palomo; Rosa L. Segura; Gloria Fernandez-Lorente; José Manuel Guisán; Roberto Fernandez-Lafuente; Enzymatic resolution of (±)-glycidyl butyrate in aqueous media. Strong modulation of the properties of the lipase from Rhizopus oryzae via immobilization techniques. Tetrahedron: Asymmetry 2004, 15, 1157-1161, 10.1016/j.tetasy.2004.03.003.
- Rahime Songür; Binnaz Lurçi; Emine Bayraktar; Ülkü Mehmetoğlu; Ayhan S. Demir; Enantioselective Production of Benzoin from Benzoin Acetate via Kinetic Resolution and Deracemization usingRhizopus oryzae. Artificial Cells, Blood Substitutes, and Biotechnology 2010, 39, 162-168, 10.3109/10731199.2010.516261.
- Zaida Cabrera; Jose M. Palomo; Enantioselective desymmetrization of prochiral diesters catalyzed by immobilized Rhizopus oryzae lipase. Tetrahedron: Asymmetry 2011, 22, 2080-2084, 10.1016/j.tetasy.2011.11.012.
- Maryam Yousefi; Mehdi Mohammadi; Zohreh Habibi; Enantioselective resolution of racemic ibuprofen esters using different lipases immobilized on octyl sepharose. Journal of Molecular Catalysis B: Enzymatic 2014, 104, 87-94, 10.1016/j.molcatb.2014.03.005.
- T. Matsumoto; M. Ito; H. Fukuda; Akihiko Kondo; Enantioselective transesterification using lipase-displaying yeast whole-cell biocatalyst. Applied Microbiology and Biotechnology 2004, 64, 481-485, 10.1007/s00253-003-1486-1.
- Yurie Nakamura; Takeshi Matsumoto; Fumiki Nomoto; Mitsuyoshi Ueda; Hideki Fukuda; Akihiko Kondo; Enhancement of Activity of Lipase-Displaying Yeast Cells and Their Application to Optical Resolution of (R,S)-1-Benzyloxy-3-Chloro-2-Propyl Monosuccinate. Biotechnology Progress 2006, 22, 998-1002, 10.1021/bp060136m.
- Zaida Cabrera; Jose M. Palomo; Enantioselective desymmetrization of prochiral diesters catalyzed by immobilized Rhizopus oryzae lipase. Tetrahedron: Asymmetry 2011, 22, 2080-2084, 10.1016/j.tetasy.2011.11.012.
- Maryam Yousefi; Mehdi Mohammadi; Zohreh Habibi; Enantioselective resolution of racemic ibuprofen esters using different lipases immobilized on octyl sepharose. Journal of Molecular Catalysis B: Enzymatic 2014, 104, 87-94, 10.1016/j.molcatb.2014.03.005.
- T. Matsumoto; M. Ito; H. Fukuda; Akihiko Kondo; Enantioselective transesterification using lipase-displaying yeast whole-cell biocatalyst. Applied Microbiology and Biotechnology 2004, 64, 481-485, 10.1007/s00253-003-1486-1.
- Yurie Nakamura; Takeshi Matsumoto; Fumiki Nomoto; Mitsuyoshi Ueda; Hideki Fukuda; Akihiko Kondo; Enhancement of Activity of Lipase-Displaying Yeast Cells and Their Application to Optical Resolution of (R,S)-1-Benzyloxy-3-Chloro-2-Propyl Monosuccinate. Biotechnology Progress 2006, 22, 998-1002, 10.1021/bp060136m.




