
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Izabela Waśko | -- | 12434 | 2022-11-14 13:47:59 | | | |
| 2 | Beatrix Zheng | -6812 word(s) | 5622 | 2022-11-16 10:54:04 | | | | |
| 3 | Beatrix Zheng | -17 word(s) | 5605 | 2022-11-16 11:00:57 | | | | |
| 4 | Beatrix Zheng | Meta information modification | 5605 | 2022-11-17 11:02:56 | | | | |
| 5 | Beatrix Zheng | Meta information modification | 5605 | 2022-11-17 11:06:52 | | |
Video Upload Options
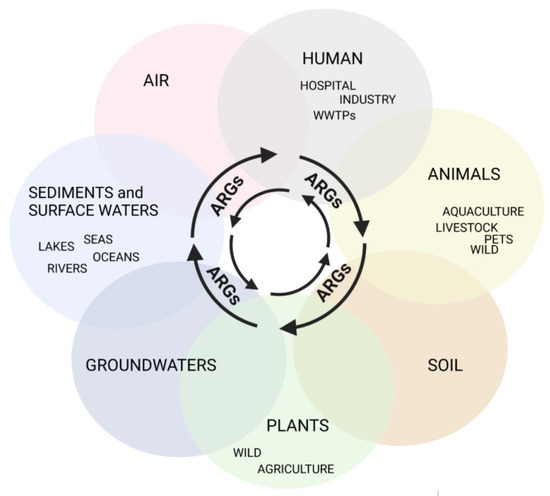
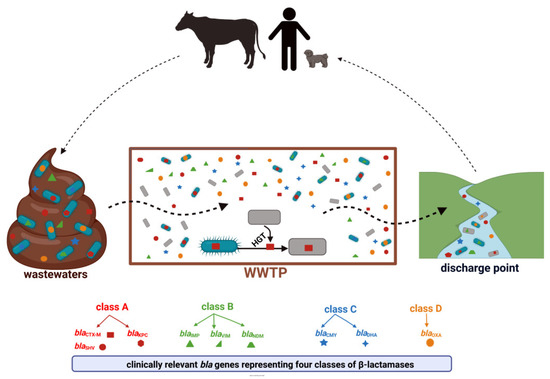
Antimicrobial resistance (AMR) is one of the largest global concerns due to its influence in multiple areas, which is consistent with One Health’s concept of close interconnections between people, animals, plants, and their shared environments. Antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs) circulate constantly in various niches, sediments, water sources, soil, and wastes of the animal and plant sectors, and is linked to human activities. Sewage of different origins gets to the wastewater treatment plants (WWTPs), where ARB and ARG removal efficiency is still insufficient, leading to their transmission to discharge points and further dissemination. Thus, WWTPs are believed to be reservoirs of ARGs and the source of spreading AMR. According to a World Health Organization report, the most critical pathogens for public health include Gram-negative bacteria resistant to third-generation cephalosporins and carbapenems (last-choice drugs), which represent β-lactams, the most widely used antibiotics. Therefore, this research presents the available research data for ARGs in WWTPs that confer resistance to β-lactam antibiotics, with a particular emphasis on clinically important life-threatening mechanisms of resistance, including extended-spectrum β-lactamases (ESBLs) and carbapenemases (KPC, NDM).
1. Introduction
2. Clinically Significant β-Lactam Resistance Genes in Wastewater Treatment Plants—The Occurrence and Distribution
2.1. Class A β-Lactamases
2.1.1. Class A β-Lactamases—Occurrence and Variability in WWTPs-Linked Samples
2.1.2. Class A β-Lactamases—Removal during the Treatment Process
2.2. Class B β-Lactamases
2.2.1. IMP and VIM β-Lactamases in WWTPs-Linked Samples
2.2.2. New Delhi Metallo-β-Lactamase (NDM) in WWTPs-Linked Samples
2.3. Class C β-Lactamases
2.3.1. Class C β-Lactamases in WWTPs-Linked Samples
2.4. Class D β-Lactamases
2.4.1. OXA Family β-Lactamases Carried in ARB
2.4.2. OXA Family β-Lactamases in Direct WWTP Samples—Occurrence and Removal
3. Conclusions
References
- WHO. Ten Threats to Global Health in 2019; World Health Organization: Geneva, Switzerland, 2019; pp. 1–18. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 10 May 2022).
- CDC. Antibiotic Resistance Threats in the United States 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 1 April 2022).
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327.
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655.
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month 2020, 66, 100971.
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281.
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 1–20.
- Alexander, J.; Hembach, N.; Schwartz, T. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 2020, 10, 8952.
- Vaz-Moreira, I.; Harnisz, M.; Abreu-Silva, J.; Rolbiecki, D.; Korzeniewska, E.; Luczkiewicz, A.; Manaia, C.M.; Plaza, G. Antibiotic resistance in wastewater, does the context matter? Poland and Portugal as a case study. Crit. Rev. Environ. Sci. Technol. 2022, 52, 4194–4216.
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964.
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023.
- Wang, J.; Chu, L.; Wojnárovits, L.; Takács, E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview. Sci. Total Environ. 2020, 744, 140997.
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment—Occurrence and environmental implications. Eur. J. Pharm. 2020, 866, 172813.
- Uluseker, C.; Kaster, K.M.; Thorsen, K.; Basiry, D.; Shobana, S.; Jain, M.; Kumar, G.; Kommedal, R.; Pala-Ozkok, I. A Review on Occurrence and Spread of Antibiotic Resistance in Wastewaters and in Wastewater Treatment Plants: Mechanisms and Perspectives. Front. Microbiol. 2021, 12, 717809.
- Zieliński, W.; Korzeniewska, E.; Harnisz, M.; Drzymała, J.; Felis, E.; Bajkacz, S. Wastewater treatment plants as a reservoir of integrase and antibiotic resistance genes—An epidemiological threat to workers and environment. Environ. Int. 2021, 156, 106641.
- Miłobedzka, A.; Ferreira, C.; Vaz-Moreira, I.; Calderón-Franco, D.; Gorecki, A.; Purkrtova, S.; Bartacek, J.; Dziewit, L.; Singleton, C.M.; Nielsen, P.H.; et al. Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the One-Health cycle. J. Hazard. Mater. 2022, 424, 127407.
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721.
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173.
- Czekalski, N.; Gascón Díez, E.; Bürgmann, H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 2014, 8, 1381–1390.
- Kotlarska, E.; Łuczkiewicz, A.; Pisowacka, M.; Burzyński, A. Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. 2015, 22, 2018–2030.
- Chaturvedi, P.; Chaurasia, D.; Pandey, A.; Gupta, P. Co-occurrence of multidrug resistance, β-lactamase and plasmid mediated AmpC genes in bacteria isolated from river Ganga, northern India. Environ. Pollut. 2020, 267, 115502.
- Griffin, D.W.; Banks, K.; Gregg, K.; Shedler, S.; Walker, B.K. Antibiotic Resistance in Marine Microbial Communities Proximal to a Florida Sewage Outfall System. Antibiotics 2020, 9, 118.
- Kvesić, M.; Kalinić, H.; Dželalija, M.; Šamanić, I.; Andričević, R.; Maravić, A. Microbiome and antibiotic resistance profiling in submarine effluent-receiving coastal waters in Croatia. Environ. Pollut. 2022, 292, 118282.
- Sabri, N.A.; Schmitt, H.; Van Der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245.
- Rodríguez-Molina, D.; Berglund, F.; Blaak, H.; Flach, C.-F.; Kemper, M.; Marutescu, L.; Gradisteanu, G.P.; Popa, M.; Spießberger, B.; Weinmann, T.; et al. Carriage of ESBL-producing Enterobacterales in wastewater treatment plant workers and surrounding residents—The AWARE Study. Eur. J. Clin. Microbiol. Infect. Dis. 2021.
- Leonard, A.F.; Morris, D.; Schmitt, H.; Gaze, W.H. Natural recreational waters and the risk that exposure to antibiotic resistant bacteria poses to human health. Curr. Opin. Microbiol. 2022, 65, 40–46.
- Stanton, I.C.; Bethel, A.; Leonard, A.F.C.; Gaze, W.H.; Garside, R. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: A systematic map. Environ. Evid. 2022, 11, 1–24.
- Zhang, X.-X.; Zhang, T.; Fang, H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414.
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360.
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124.
- Pärnänen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124.
- Swami, O.C. Strategies to Combat Antimicrobial Resistance. J. Clin. Diagn. Res. 2014, 8, 8–11.
- WHO. Global Action Plan on Antimicrobial Resistance 2015. Available online: https://apps.who.int/iris/handle/10665/193736 (accessed on 10 May 2022).
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317.
- Barancheshme, F.; Munir, M. Strategies to Combat Antibiotic Resistance in the Wastewater Treatment Plants. Front. Microbiol. 2018, 8, 2603.
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380.
- Marano, R.B.M.; Fernandes, T.; Manaia, C.M.; Nunes, O.; Morrison, D.; Berendonk, T.U.; Kreuzinger, N.; Tenson, T.; Corno, G.; Fatta-Kassinos, D.; et al. A global multinational survey of cefotaxime-resistant coliforms in urban wastewater treatment plants. Environ. Int. 2020, 144, 106035.
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269.
- Daoud, Z.; Rolain, J.-M. Editorial: “One Health” Approach for Revealing Reservoirs and Transmission of Antimicrobial Resistance. Front. Microbiol. 2022, 12, 2021–2023.
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402.
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 2020, 8, 13.
- Baraniak, A.; Fiett, J.; Mrówka, A.; Walory, J.; Hryniewicz, W.; Gniadkowski, M. Evolution of TEM-Type Extended-Spectrum β-Lactamases in Clinical Enterobacteriaceae Strains in Poland. Antimicrob. Agents Chemother. 2005, 49, 1872–1880.
- De Champs, C.; Sirot, D.; Chanal, C.; Bonnet, R.; Sirot, J. A 1998 Survey of Extended-Spectrum β-Lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 2000, 44, 3177–3179.
- Livermore, D.M.; Canton, R.; Gniadkowski, M.; Nordmann, P.; Rossolini, G.M.; Arlet, G.; Ayala, J.; Coque, T.M.; Kern-Zdanowicz, I.; Luzzaro, F.; et al. CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2006, 59, 165–174.
- Doi, Y.; Iovleva, A.; Bonomo, R.A. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J. Travel Med. 2017, 24, S44–S51.
- Wiener, J. Multiple Antibiotic–Resistant Klebsiella and Escherichia coli in Nursing Homes. JAMA 1999, 281, 517.
- Pitout, J.D.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005, 56, 52–59.
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686.
- Nordmann, P.; Cuzon, G.; Naas, T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009, 9, 228–236.
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798.
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796.
- Li, L.; Nesme, J.; Quintela-Baluja, M.; Balboa, S.; Hashsham, S.; Williams, M.R.; Yu, Z.; Sørensen, S.J.; Graham, D.W.; Romalde, J.L.; et al. Extended-Spectrum β-Lactamase and Carbapenemase Genes are Substantially and Sequentially Reduced during Conveyance and Treatment of Urban Sewage. Environ. Sci. Technol. 2021, 55, 5939–5949.
- Zaatout, N.; Bouras, S.; Slimani, N. Prevalence of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in wastewater: A systematic review and meta-analysis. J. Water Health 2021, 19, 705–723.
- Gündoğdu, A.; Jennison, A.V.; Smith, H.V.; Stratton, H.; Katouli, M. Extended-spectrum β-lactamase producing Escherichia coli in hospital wastewaters and sewage treatment plants in Queensland, Australia. Can. J. Microbiol. 2013, 59, 737–745.
- Korzeniewska, E.; Harnisz, M. Extended-spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. J. Environ. Manag. 2013, 128, 904–911.
- Makowska, N.; Philips, A.; Dabert, M.; Nowis, K.; Trzebny, A.; Koczura, R.; Mokracka, J. Metagenomic analysis of β-lactamase and carbapenemase genes in the wastewater resistome. Water Res. 2020, 170, 115277.
- Yuan, W.; Tian, T.; Yang, Q.; Riaz, L. Transfer potentials of antibiotic resistance genes in Escherichia spp. strains from different sources. Chemosphere 2020, 246, 125736.
- Ali, A.; Sultan, I.; Mondal, A.H.; Siddiqui, M.T.; Gogry, F.A.; Haq, Q.M.R. Lentic and effluent water of Delhi-NCR: A reservoir of multidrug-resistant bacteria harbouring bla CTX-M, bla TEM and bla SHV type ESBL genes. J. Water Health 2021, 19, 592–603.
- Fadare, F.T.; Okoh, A.I. The Abundance of Genes Encoding ESBL, pAmpC and Non-β-Lactam Resistance in Multidrug-Resistant Enterobacteriaceae Recovered from Wastewater Effluents. Front. Environ. Sci. 2021, 9, 1–12.
- Gumede, S.N.; Abia, A.L.K.; Amoako, D.G.; Essack, S.Y. Analysis of Wastewater Reveals the Spread of Diverse Extended-Spectrum β-Lactamase-Producing E. coli Strains in uMgungundlovu District, South Africa. Antibiotics 2021, 10, 860.
- Alouache, S.; Estepa, V.; Messai, Y.; Ruiz, E.; Torres, C.; Bakour, R. Characterization of ESBLs and Associated Quinolone Resistance in Escherichia coli and Klebsiella pneumoniae Isolates from an Urban Wastewater Treatment Plant in Algeria. Microb. Drug Resist. 2014, 20, 30–38.
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater treatment plants release large amounts of extended-spectrum β-lactamase-producing escherichia coli into the environment. Clin. Infect. Dis. 2014, 58, 1658–1665.
- Ojer-Usoz, E.; González, D.; García-Jalón, I.; Vitas, A.I. High dissemination of extended-spectrum β-lactamase-producing Enterobacteriaceae in effluents from wastewater treatment plants. Water Res. 2014, 56, 37–47.
- Akiba, M.; Sekizuka, T.; Yamashita, A.; Kuroda, M.; Fujii, Y.; Murata, M.; Lee, K.I.; Joshua, D.I.; Balakrishna, K.; Bairy, I.; et al. Distribution and relationships of antimicrobial resistance determinants among extended-spectrum-cephalosporin-resistant or carbapenem-resistant Escherichia coli isolates from rivers and sewage treatment plants in India. Antimicrob. Agents Chemother. 2016, 60, 2972–2980.
- Caltagirone, M.; Nucleo, E.; Spalla, M.; Zara, F.; Novazzi, F.; Marchetti, V.M.; Piazza, A.; Bitar, I.; De Cicco, M.; Paolucci, S.; et al. Occurrence of Extended Spectrum β-Lactamases, KPC-Type, and MCR-1.2-Producing Enterobacteriaceae from Wells, River Water, and Wastewater Treatment Plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 2017, 8, 1–12.
- Conte, D.; Palmeiro, J.K.; da Silva Nogueira, K.; de Lima, T.M.R.; Cardoso, M.A.; Pontarolo, R.; Degaut Pontes, F.L.; Dalla-Costa, L.M. Characterization of CTX-M enzymes, quinolone resistance determinants, and antimicrobial residues from hospital sewage, wastewater treatment plant, and river water. Ecotoxicol. Environ. Saf. 2017, 136, 62–69.
- Guyomard-Rabenirina, S.; Dartron, C.; Falord, M.; Sadikalay, S.; Ducat, C.; Richard, V.; Breurec, S.; Gros, O.; Talarmin, A. Resistance to antimicrobial drugs in different surface waters and wastewaters of Guadeloupe. PLoS ONE 2017, 12, e0173155.
- Galler, H.; Feierl, G.; Petternel, C.; Reinthaler, F.; Haas, D.; Habib, J.; Kittinger, C.; Luxner, J.; Zarfel, G. Multiresistant Multiresistant Bacteria Isolated from Activated Sludge in Austria. Int. J. Environ. Res. Public Health 2018, 15, 479.
- Yang, Y.; Zhang, T.; Zhang, X.-X.; Liang, D.-W.; Zhang, M.; Gao, D.-W.; Zhu, H.-G.; Huang, Q.-G.; Fang, H.H.P. Quantification and characterization of β-lactam resistance genes in 15 sewage treatment plants from East Asia and North America. Appl. Microbiol. Biotechnol. 2012, 95, 1351–1358.
- Khan, M.A.; Thurgood, N.E.; Faheem, S.M.; Rais, N.; Ansari, M.Z.; Kaleem, S.M.; Khan, S.T. Occurrence of Extended Spectrum Beta-Lactamase Gram-Negative Bacteria from Non-Clinical Sources in Dubai, United Arab Emirates. Water 2020, 12, 2562.
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Felis, E.; Bajkacz, S.; Jachimowicz, P.; Niestępski, S.; Konopka, I. Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater. 2020, 381, 121221.
- Surleac, M.; Barbu, I.C.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M.; et al. Whole genome sequencing snapshot of multidrug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079.
- Smyth, C.; O’Flaherty, A.; Walsh, F.; Do, T.T. Antibiotic resistant and extended-spectrum β-lactamase producing faecal coliforms in wastewater treatment plant effluent. Environ. Pollut. 2020, 262, 114244.
- Aristizábal-Hoyos, A.M.; Rodríguez, E.A.; Arias, L.; Jiménez, J.N. High clonal diversity of multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in a wastewater treatment plant. J. Environ. Manag. 2019, 245, 37–47.
- Amador, P.P.; Fernandes, R.M.; Prudêncio, M.C.; Barreto, M.P.; Duarte, I.M. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of Class A and Class C β-lactamases. J. Environ. Sci. Health Part A 2015, 50, 26–39.
- Proia, L.; Anzil, A.; Borrego, C.; Farrè, M.; Llorca, M.; Sanchis, J.; Bogaerts, P.; Balcázar, J.L.; Servais, P. Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. J. Hazard. Mater. 2018, 358, 33–43.
- Zhang, A.; Call, D.R.; Besser, T.E.; Liu, J.; Jones, L.; Wang, H.; Davis, M.A. β-lactam resistance genes in bacteriophage and bacterial DNA from wastewater, river water, and irrigation water in Washington State. Water Res. 2019, 161, 335–340.
- Adegoke, A.A.; Madu, C.E.; Aiyegoro, O.A.; Stenström, T.A.; Okoh, A.I. Antibiogram and beta-lactamase genes among cefotaxime resistant E. coli from wastewater treatment plant. Antimicrob. Resist. Infect. Control 2020, 9, 46.
- Piotrowska, M.; Kowalska, S.; Popowska, M. Diversity of β-lactam resistance genes in gram-negative rods isolated from a municipal wastewater treatment plant. Ann. Microbiol. 2019, 69, 591–601.
- Hubeny, J.; Ciesielski, S.; Harnisz, M.; Korzeniewska, E.; Dulski, T.; Jałowiecki, Ł.; Płaza, G. Impact of Hospital Wastewater on the Occurrence and Diversity of Beta-Lactamase Genes During Wastewater Treatment with an Emphasis on Carbapenemase Genes: A Metagenomic Approach. Front. Environ. Sci. 2021, 9, 738158.
- Teban-Man, A.; Farkas, A.; Baricz, A.; Hegedus, A.; Szekeres, E.; Pârvu, M.; Coman, C. Wastewaters, with or without Hospital Contribution, Harbour MDR, Carbapenemase-Producing, but Not Hypervirulent Klebsiella pneumoniae. Antibiotics 2021, 10, 361.
- Galler, H.; Feierl, G.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Grisold, A.J.; Luxner, J.; Zarfel, G. KPC-2 and OXA-48 carbapenemase-harbouring Enterobacteriaceae detected in an Austrian wastewater treatment plant. Clin. Microbiol. Infect. 2014, 20, O132–O134.
- Bengtsson-Palme, J.; Hammarén, R.; Pal, C.; Östman, M.; Björlenius, B.; Flach, C.-F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.G.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712.
- Piotrowska, M.; Przygodzińska, D.; Matyjewicz, K.; Popowska, M. Occurrence and Variety of β-Lactamase Genes among Aeromonas spp. Isolated from Urban Wastewater Treatment Plant. Front. Microbiol. 2017, 8, 863.
- Subirats, J.; Royo, E.; Balcázar, J.L.; Borrego, C.M. Real-time PCR assays for the detection and quantification of carbapenemase genes (bla KPC, bla NDM, and bla OXA-48) in environmental samples. Environ. Sci. Pollut. Res. 2017, 24, 6710–6714.
- Zurfluh, K.; Bagutti, C.; Brodmann, P.; Alt, M.; Schulze, J.; Fanning, S.; Stephan, R.; Nüesch-Inderbinen, M. Wastewater is a reservoir for clinically relevant carbapenemase- and 16s rRNA methylase-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2017, 50, 436–440.
- Müller, H.; Sib, E.; Gajdiss, M.; Klanke, U.; Lenz-Plet, F.; Barabasch, V.; Albert, C.; Schallenberg, A.; Timm, C.; Zacharias, N.; et al. Dissemination of multi-resistant Gram-negative bacteria into German wastewater and surface waters. FEMS Microbiol. Ecol. 2018, 94, fiy057.
- Cacace, D.; Fatta-Kassinos, D.; Manaia, C.M.; Cytryn, E.; Kreuzinger, N.; Rizzo, L.; Karaolia, P.; Schwartz, T.; Alexander, J.; Merlin, C.; et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019, 162, 320–330.
- Mathys, D.A.; Mollenkopf, D.F.; Feicht, S.M.; Adams, R.J.; Albers, A.L.; Stuever, D.M.; Grooters, S.V.; Ballash, G.A.; Daniels, J.B.; Wittum, T.E. Carbapenemase-producing Enterobacteriaceae and Aeromonas spp. Present in wastewater treatment plant effluent and nearby surface waters in the US. PLoS ONE 2018, 14, e0218650.
- Hoelle, J.; Johnson, J.R.; Johnston, B.D.; Kinkle, B.; Boczek, L.; Ryu, H.; Hayes, S. Survey of US wastewater for carbapenem-resistant Enterobacteriaceae. J. Water Health 2019, 17, 219–226.
- Cooper, A.L.; Carter, C.; McLeod, H.; Wright, M.; Sritharan, P.; Tamber, S.; Wong, A.; Carrillo, C.D.; Blais, B.W. Detection of carbapenem-resistance genes in bacteria isolated from wastewater in Ontario. FACETS 2021, 6, 569–591.
- Ebomah, K.E.; Okoh, A.I. Detection of Carbapenem-Resistance Genes in Klebsiella Species Recovered from Selected Environmental Niches in the Eastern Cape Province, South Africa. Antibiotics 2020, 9, 425.
- Yang, F.; Mao, D.; Zhou, H.; Luo, Y. Prevalence and Fate of Carbapenemase Genes in a Wastewater Treatment Plant in Northern China. PLoS ONE 2016, 11, e0156383.
- Ng, C.; Tan, B.; Jiang, X.-T.; Gu, X.; Chen, H.; Schmitz, B.W.; Haller, L.; Charles, F.R.; Zhang, T.; Gin, K. Metagenomic and Resistome Analysis of a Full-Scale Municipal Wastewater Treatment Plant in Singapore Containing Membrane Bioreactors. Front. Microbiol. 2019, 10, 172.
- Ng, C.; Tay, M.; Tan, B.; Le, T.-H.; Haller, L.; Chen, H.; Koh, T.H.; Barkham, T.M.S.; Thompson, J.R.; Gin, K.Y.H. Characterization of Metagenomes in Urban Aquatic Compartments Reveals High Prevalence of Clinically Relevant Antibiotic Resistance Genes in Wastewaters. Front. Microbiol. 2017, 8, 2200.
- Khan, F.A.; Söderquist, B.; Jass, J. Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater. Front. Microbiol. 2019, 10, 688.
- Araújo, S.; Sousa, M.; Tacão, M.; Baraúna, R.A.; Silva, A.; Ramos, R.; Alves, A.; Manaia, C.M.; Henriques, I. Carbapenem-resistant bacteria over a wastewater treatment process: Carbapenem-resistant Enterobacteriaceae in untreated wastewater and intrinsically-resistant bacteria in final effluent. Sci. Total Environ. 2021, 782, 146892.
- Zhang, D.; Peng, Y.; Chan, C.-L.; On, H.; Wai, H.K.-F.; Shekhawat, S.S.; Gupta, A.B.; Varshney, A.K.; Chuanchuen, R.; Zhou, X.; et al. Metagenomic Survey Reveals More Diverse and Abundant Antibiotic Resistance Genes in Municipal Wastewater Than Hospital Wastewater. Front. Microbiol. 2021, 12, 712843.
- Reinthaler, F.F.; Feierl, G.; Galler, H.; Haas, D.; Leitner, E.; Mascher, F.; Melkes, A.; Posch, J.; Winter, I.; Zarfel, G. ESBL-producing E. coli in Austrian sewage sludge. Water Res. 2010, 44, 1981–1985.
- Dolejska, M.; Frolkova, P.; Florek, M.; Jamborova, I.; Purgertova, M.; Kutilova, I.; Cizek, A.; Guenther, S.; Literak, I. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J. Antimicrob. Chemother. 2011, 66, 2784–2790.
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102.
- Čornejová, T.; Venglovsky, J.; Gregova, G.; Kmetova, M.; Kmet, V. Extended spectrum beta-lactamases in Escherichia coli from municipal wastewater. Ann. Agric. Environ. Med. 2015, 22, 447–450.
- Kwak, Y.-K.; Colque, P.; Byfors, S.; Giske, C.G.; Möllby, R.; Kühn, I. Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: Does it reflect the resistance trends in the society? Int. J. Antimicrob. Agents 2015, 45, 25–32.
- Dropa, M.; Lincopan, N.; Balsalobre, L.C.; Oliveira, D.E.; Moura, R.A.; Fernandes, M.R.; da Silva, Q.M.; Matté, G.R.; Sato, M.I.Z.; Matté, M.H. Genetic background of novel sequence types of CTX-M-8- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae from public wastewater treatment plants in São Paulo, Brazil. Environ. Sci. Pollut. Res. 2016, 23, 4953–4958.
- Silva, I.; Tacão, M.; Tavares, R.D.S.; Miranda, R.; Araújo, S.; Manaia, C.M.; Henriques, I. Fate of cefotaxime-resistant Enterobacteriaceae and ESBL-producers over a full-scale wastewater treatment process with UV disinfection. Sci. Total Environ. 2018, 639, 1028–1037.
- Haberecht, H.B.; Nealon, N.J.; Gilliland, J.R.; Holder, A.V.; Runyan, C.; Oppel, R.C.; Ibrahim, H.M.; Mueller, L.; Schrupp, F.; Vilchez, S.; et al. Antimicrobial-Resistant Escherichia coli from Environmental Waters in Northern Colorado. J. Environ. Public Health 2019, 2019, 1–13.
- Raven, K.E.; Ludden, C.; Gouliouris, T.; Blane, B.; Naydenova, P.; Brown, N.M.; Parkhill, J.; Peacock, S.J. Genomic surveillance of Escherichia coli in municipal wastewater treatment plants as an indicator of clinically relevant pathogens and their resistance genes. Microb. Genom. 2019, 5, e000267.
- Sghaier, S.; Abbassi, M.S.; Pascual, A.; Serrano, L.; Díaz-De-Alba, P.; Said, M.B.; Hassen, B.; Ibrahim, C.; Hassen, A.; López-Cerero, L. Extended-spectrum β-lactamase-producing Enterobacteriaceae from animal origin and wastewater in Tunisia: First detection of O25b-B23-CTX-M-27-ST131 Escherichia coli and CTX-M-15/OXA-204-producing Citrobacter freundii from wastewater. J. Glob. Antimicrob. Resist. 2019, 17, 189–194.
- Tanaka, H.; Hayashi, W.; Iimura, M.; Taniguchi, Y.; Soga, E.; Matsuo, N.; Kawamura, K.; Arakawa, Y.; Nagano, Y.; Nagano, N. Wastewater as a Probable Environmental Reservoir of Extended-Spectrum-β-Lactamase Genes: Detection of Chimeric β-Lactamases CTX-M-64 and CTX-M-123. Appl. Environ. Microbiol. 2019, 85, 1–12.
- Schages, L.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. Winter is coming—Impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant. Sci. Total Environ. 2020, 712, 136499.
- Urano, N.; Okai, M.; Tashiro, Y.; Takeuchi, A.; Endo, R.; Ishida, M.; Takashio, M. Behavior of Antibiotic-Resistant Fecal Coliforms in the Stream of a Sewage Treatment Plant in Tokyo. Adv. Microbiol. 2020, 10, 318–330.
- Liedhegner, E.; Bojar, B.; Beattie, R.E.; Cahak, C.; Hristova, K.R.; Skwor, T. Similarities in Virulence and Extended Spectrum Beta-Lactamase Gene Profiles among Cefotaxime-Resistant Escherichia coli Wastewater and Clinical Isolates. Antibiotics 2022, 11, 260.
- Igbinosa, I.H.; Okoh, A.I. Antibiotic Susceptibility Profile of Aeromonas Species Isolated from Wastewater Treatment Plant. Sci. World J. 2012, 2012, 1–6.
- Ben Said, L.; Jouini, A.; Alonso, C.A.; Klibi, N.; Dziri, R.; Boudabous, A.; Ben Slama, K.; Torres, C. Characteristics of extended-spectrum β-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci. Total Environ. 2016, 550, 1103–1109.
- Ludden, C.; Reuter, S.; Judge, K.; Gouliouris, T.; Blane, B.; Coll, F.; Naydenova, P.; Hunt, M.; Tracey, A.; Hopkins, K.L.; et al. Sharing of carbapenemase-encoding plasmids between Enterobacteriaceae in UK sewage uncovered by MinION sequencing. Microb. Genom. 2017, 3, e000114.
- Zhang, S.; Han, B.; Gu, J.; Wang, C.; Wang, P.; Ma, Y.; Cao, J.; He, Z. Fate of antibiotic resistant cultivable heterotrophic bacteria and antibiotic resistance genes in wastewater treatment processes. Chemosphere 2015, 135, 138–145.
- Nzima, B.; Adegoke, A.A.; Ofon, U.A.; Al-Dahmoshi, H.O.M.; Saki, M.; Ndubuisi-Nnaji, U.U.; Inyang, C.U. Resistotyping and extended-spectrum beta-lactamase genes among Escherichia coli from wastewater treatment plants and recipient surface water for reuse in South Africa. New Microbes New Infect. 2020, 38, 100803.
- Gomi, R.; Matsuda, T.; Matsumura, Y.; Yamamoto, M.; Tanaka, M.; Ichiyama, S.; Yoneda, M. Occurrence of clinically important lineages, including the sequence type 131 C1-M27 subclone, among extended-spectrum--lactamase-producing Escherichia coli in wastewater. Antimicrob. Agents Chemother. 2017, 61, e00564-17.
- Finn, T.J.; Scriver, L.; Lam, L.; Duong, M.; Peirano, G.; Lynch, T.; Dong, T.; Pitout, J.D.D.; DeVinney, R. A comprehensive account of escherichia coli sequence type 131 in wastewater reveals an abundance of fluoroquinolone-resistant clade a strains. Appl. Environ. Microbiol. 2020, 86, 1–11.
- Zhi, S.; Stothard, P.; Banting, G.; Scott, C.; Huntley, K.; Ryu, K.; Otto, S.; Ashbolt, N.; Checkley, S.; Dong, T.; et al. Characterization of water treatment-resistant and multidrug-resistant urinary pathogenic Escherichia coli in treated wastewater. Water Res. 2020, 182, 115827.
- Mesquita, E.; Ribeiro, R.; Silva, C.J.C.; Alves, R.; Baptista, R.; Condinho, S.; Rosa, M.J.; Perdigão, J.; Caneiras, C.; Duarte, A. An update on wastewater multi-resistant bacteria: Identification of clinical pathogens such as escherichia coli o25b:H4-b2-st131-producing ctx-m-15 esbl and kpc-3 carbapenemase-producing klebsiella oxytoca. Microorganisms 2021, 9, 576.
- Hembach, N.; Schmid, F.; Alexander, J.; Hiller, C.; Rogall, E.T.; Schwartz, T. Occurrence of the mcr-1 Colistin Resistance Gene and other Clinically Relevant Antibiotic Resistance Genes in Microbial Populations at Different Municipal Wastewater Treatment Plants in Germany. Front. Microbiol. 2017, 8, 1282.
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.-H.; Gützkow, T.; Eichler, W.; Pühler, A.; Schlüter, A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 2009, 155, 2306–2319.
- Wen, Q.; Yang, L.; Duan, R.; Chen, Z. Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin, Northeast China. Environ. Pollut. 2016, 212, 34–40.
- Narciso-da-Rocha, C.; Rocha, J.; Vaz-Moreira, I.; Lira, F.; Tamames, J.; Henriques, I.; Martinez, J.L.; Manaia, C.M. Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environ. Int. 2018, 118, 179–188.
- Neudorf, K.D.; Huang, Y.N.; Ragush, C.M.; Yost, C.K.; Jamieson, R.C.; Truelstrup Hansen, L. Antibiotic resistance genes in municipal wastewater treatment systems and receiving waters in Arctic Canada. Sci. Total Environ. 2017, 598, 1085–1094.
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242.
- Laht, M.; Karkman, A.; Voolaid, V.; Ritz, C.; Tenson, T.; Virta, M.; Kisand, V. Abundances of Tetracycline, Sulphonamide and Beta-Lactam Antibiotic Resistance Genes in Conventional Wastewater Treatment Plants (WWTPs) with Different Waste Load. PLoS ONE 2014, 9, e103705.
- Rafraf, I.D.; Lekunberri, I.; Sànchez-Melsió, A.; Aouni, M.; Borrego, C.M.; Balcázar, J.L. Abundance of antibiotic resistance genes in five municipal wastewater treatment plants in the Monastir Governorate, Tunisia. Environ. Pollut. 2016, 219, 353–358.
- Kowalski, M.; Wolany, J.; Pastuszka, J.S.; Płaza, G.; Wlazło, A.; Ulfig, K.; Malina, A. Characteristics of airborne bacteria and fungi in some Polish wastewater treatment plants. Int. J. Environ. Sci. Technol. 2017, 14, 2181–2192.
- Ginn, O.; Rocha-Melogno, L.; Bivins, A.; Lowry, S.; Cardelino, M.; Nichols, D.; Tripathi, S.N.; Soria, F.; Andrade, M.; Bergin, M.; et al. Detection and Quantification of Enteric Pathogens in Aerosols near Open Wastewater Canals in Cities with Poor Sanitation. Environ. Sci. Technol. 2021, 55, 14758–14771.
- Wengenroth, L.; Berglund, F.; Blaak, H.; Chifiriuc, M.C.; Flach, C.-F.; Pircalabioru, G.G.; Larsson, D.G.J.; Marutescu, L.; van Passel, M.W.J.; Popa, M.; et al. Antibiotic Resistance in Wastewater Treatment Plants and Transmission Risks for Employees and Residents: The Concept of the AWARE Study. Antibiotics 2021, 10, 478.
- Lee, G.; Yoo, K. A review of the emergence of antibiotic resistance in bioaerosols and its monitoring methods. Rev. Environ. Sci. Bio/Technol. 2022, 21, 799–827.
- Gaviria-Figueroa, A.; Preisner, E.C.; Hoque, S.; Feigley, C.E.; Norman, R.S. Emission and dispersal of antibiotic resistance genes through bioaerosols generated during the treatment of municipal sewage. Sci. Total Environ. 2019, 686, 402–412.
- Palzkill, T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104.
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101.
- Cui, X.; Zhang, H.; Du, H. Carbapenemases in Enterobacteriaceae: Detection and Antimicrobial Therapy. Front. Microbiol. 2019, 10, 1823.
- Kazmierczak, K.M.; Rabine, S.; Hackel, M.; McLaughlin, R.E.; Biedenbach, D.J.; Bouchillon, S.K.; Sahm, D.F.; Bradford, P.A. Multiyear, Multinational Survey of the Incidence and Global Distribution of Metallo-β-Lactamase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 1067–1078.
- Matsumura, Y.; Peirano, G.; Motyl, M.R.; Adams, M.D.; Chen, L.; Kreiswirth, B.; DeVinney, R.; Pitout, J.D.D. Global Molecular Epidemiology of IMP-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2017, 61, e02729-16.
- Matsumura, Y.; Peirano, G.; Bradford, P.A.; Motyl, M.R.; DeVinney, R.; Pitout, J.D.D. Genomic characterization of IMP and VIM carbapenemase-encoding transferable plasmids of Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 3034–3038.
- Ghaith, D.M.; Zafer, M.M.; Ismail, D.K.; Al-Agamy, M.H.; Bohol, M.F.F.; Al-Qahtani, A.; Al-Ahdal, M.N.; Elnagdy, S.M.; Mostafa, I.Y. First reported nosocomial outbreak of Serratia marcescens harboring blaIMP-4 and blaVIM-2 in a neonatal intensive care unit in Cairo, Egypt. Infect. Drug Resist. 2018, 11, 2211–2217.
- Castanheira, M.; Deshpande, L.M.; Mendes, R.E.; Canton, R.; Sader, H.S.; Jones, R.N. Variations in the Occurrence of Resistance Phenotypes and Carbapenemase Genes Among Enterobacteriaceae Isolates in 20 Years of the SENTRY Antimicrobial Surveillance Program. Open Forum Infect. Dis. 2019, 6, S23–S33.
- Pournaras, S. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-beta-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 2003, 51, 1409–1414.
- Lagatolla, C.; Edalucci, E.; Dolzani, L.; Riccio, M.L.; De Luca, F.; Medessi, E.; Rossolini, G.M.; Tonin, E.A. Molecular Evolution of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa in a Nosocomial Setting of High-Level Endemicity. J. Clin. Microbiol. 2006, 44, 2348–2353.
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054.
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, 1–37.
- van Loon, K.; Voor in ‘t holt, A.F.; Vos, M.C. A Systematic Review and Meta-analyses of the Clinical Epidemiology of Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, 1–18.
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18.
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92.
- Alexander, J.; Bollmann, A.; Seitz, W.; Schwartz, T. Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci. Total Environ. 2015, 512–513, 316–325.
- Hubeny, J.; Korzeniewska, E.; Buta-Hubeny, M.; Zieliński, W.; Rolbiecki, D.; Harnisz, M. Characterization of carbapenem resistance in environmental samples and Acinetobacter spp. isolates from wastewater and river water in Poland. Sci. Total Environ. 2022, 822, 153437.
- Khan, F.A.; Hellmark, B.; Ehricht, R.; Söderquist, B.; Jass, J. Related carbapenemase-producing Klebsiella isolates detected in both a hospital and associated aquatic environment in Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2241–2251.
- Mills, M.C.; Lee, J. The threat of carbapenem-resistant bacteria in the environment: Evidence of widespread contamination of reservoirs at a global scale. Environ. Pollut. 2019, 255, 113143.
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.-M. Carbapenemase-producing Gram-negative bacteria in aquatic environments: A review. J. Glob. Antimicrob. Resist. 2021, 25, 287–309.
- Ranjan, R.; Thatikonda, S. β-Lactam Resistance Gene NDM-1 in the Aquatic Environment: A Review. Curr. Microbiol. 2021, 78, 3634–3643.
- Lamba, M.; Graham, D.W.; Ahammad, S.Z. Hospital Wastewater Releases of Carbapenem-Resistance Pathogens and Genes in Urban India. Environ. Sci. Technol. 2017, 51, 13906–13912.
- Parvez, S.; Khan, A.U. Hospital sewage water: A reservoir for variants of New Delhi metallo-β-lactamase (NDM)- and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2018, 51, 82–88.
- Bardhan, T.; Chakraborty, M.; Bhattacharjee, B. Prevalence of colistin-resistant, carbapenem-hydrolyzing proteobacteria in hospital water bodies and out-falls of West Bengal, India. Int. J. Environ. Res. Public Health 2020, 17, 1007.
- Hwang, S.H.; Kim, Y.J. Meropenem-resistant bacteria in hospital effluents in Seoul, Korea. Environ. Monit. Assess. 2018, 190, 673.
- Haller, L.; Chen, H.; Ng, C.; Le, T.H.; Koh, T.H.; Barkham, T.; Sobsey, M.; Gin, K.Y.H. Occurrence and characteristics of extended-spectrum β-lactamase- and carbapenemase- producing bacteria from hospital effluents in Singapore. Sci. Total Environ. 2018, 615, 1119–1125.
- Suzuki, Y.; Nazareno, P.J.; Nakano, R.; Mondoy, M.; Nakano, A.; Bugayong, M.P.; Bilar, J.; Perez, M.; Medina, E.J.; Saito-Obata, M.; et al. Environmental Presence and Genetic Characteristics of Carbapenemase-Producing Enterobacteriaceae from Hospital Sewage and River Water in the Philippines. Appl. Environ. Microbiol. 2020, 86, 1–10.
- Nasri, E.; Subirats, J.; Sànchez-Melsió, A.; Ben Mansour, H.; Borrego, C.M.; Balcázar, J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017, 229, 371–374.
- Marathe, N.P.; Berglund, F.; Razavi, M.; Pal, C.; Dröge, J.; Samant, S.; Kristiansson, E.; Larsson, D.G.J. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 2019, 7, 97.
- Zhang, L.; Ma, X.; Luo, L.; Hu, N.; Duan, J.; Tang, Z.; Zhong, R.; Li, Y. The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers. Int. J. Environ. Res. Public Health 2020, 17, 1183.
- Luo, Y.; Yang, F.; Mathieu, J.; Mao, D.; Wang, Q.; Alvarez, P.J.J. Proliferation of Multidrug-Resistant New Delhi Metallo-β-lactamase Genes in Municipal Wastewater Treatment Plants in Northern China. Environ. Sci. Technol. Lett. 2014, 1, 26–30.
- Yang, F.; Mao, D.; Zhou, H.; Wang, X.; Luo, Y. Propagation of New Delhi Metallo-β-lactamase Genes (bla NDM-1) from a Wastewater Treatment Plant to Its Receiving River. Environ. Sci. Technol. Lett. 2016, 3, 138–143.
- Mantilla-Calderon, D.; Jumat, M.R.; Wang, T.; Ganesan, P.; Al-Jassim, N.; Hong, P.Y. Isolation and characterization of NDM-positive Escherichia coli from municipal wastewater in Jeddah, Saudi Arabia. Antimicrob. Agents Chemother. 2016, 60, 5223–5231.
- Stachurová, T.; Piková, H.; Bartas, M.; Semerád, J.; Svobodová, K.; Malachová, K. Beta-lactam resistance development during the treatment processes of municipal wastewater treatment plants. Chemosphere 2021, 280, 130749.
- Mahon, B.M.; Brehony, C.; McGrath, E.; Killeen, J.; Cormican, M.; Hickey, P.; Keane, S.; Hanahoe, B.; Dolan, A.; Morris, D. Indistinguishable NDM-producing Escherichia coli isolated from recreational waters, sewage, and a clinical specimen in Ireland, 2016 to 2017. Eurosurveillance 2017, 22, 30513.
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362.
- Lamba, M.; Gupta, S.; Shukla, R.; Graham, D.W.; Sreekrishnan, T.R.; Ahammad, S.Z. Carbapenem resistance exposures via wastewaters across New Delhi. Environ. Int. 2018, 119, 302–308.
- Divyashree, M.; Mani, M.K.; Shama Prakash, K.; Vijaya Kumar, D.; Veena Shetty, A.; Shetty, A.K.; Karunasagar, I. Hospital wastewater treatment reduces NDM-positive bacteria being discharged into water bodies. Water Environ. Res. 2020, 92, 562–568.
- Bonomo, R.A. β-Lactamases: A Focus on Current Challenges. Cold Spring Harb. Perspect. Med. 2017, 7, a025239.
- Philippon, A.; Arlet, G.; Jacoby, G.A. Plasmid-Determined AmpC-Type β-Lactamases. Antimicrob. Agents Chemother. 2002, 46, 1–11.
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182.
- Pérez-Etayo, L.; González, D.; Leiva, J.; Vitas, A.I. Multidrug-Resistant Bacteria Isolated from Different Aquatic Environments in the North of Spain and South of France. Microorganisms 2020, 8, 1425.
- Yim, G.; Kwong, W.; Davies, J.; Miao, V. Complex integrons containing qnrB4-ampC (bla DHA-1) in plasmids of multidrug-resistant Citrobacter freundii from wastewater. Can. J. Microbiol. 2013, 59, 110–116.
- Su, H.-C.; Ying, G.-G.; He, L.-Y.; Liu, Y.-S.; Zhang, R.-Q.; Tao, R. Antibiotic resistance, plasmid-mediated quinolone resistance (PMQR) genes and ampC gene in two typical municipal wastewater treatment plants. Environ. Sci. Process. Impacts 2014, 16, 324.
- Pitout, J.D.D.; Peirano, G.; Kock, M.M.; Strydom, K.; Matsumura, Y. The Global Ascendency of OXA-48-Type Carbapenemases. Clin. Microbiol. Rev. 2019, 33, 1–48.
- Evans, B.A.; Amyes, S.G.B. OXA β-Lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263.
- Mairi, A.; Pantel, A.; Sotto, A.; Lavigne, J.-P.; Touati, A. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 587–604.
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22.




