
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Maria L Perepechaeva | -- | 6305 | 2022-11-11 08:57:07 | | | |
| 2 | Maria L Perepechaeva | -4266 word(s) | 2039 | 2022-11-11 18:28:25 | | | | |
| 3 | Maria L Perepechaeva | Meta information modification | 2039 | 2022-11-11 18:31:08 | | | | |
| 4 | Rita Xu | Meta information modification | 2039 | 2022-11-14 02:48:26 | | | | |
| 5 | Rita Xu | Meta information modification | 2039 | 2022-11-14 08:29:55 | | |
Video Upload Options
CYP3A is an enzyme subfamily in the cytochrome P450 (CYP) superfamily and includes isoforms CYP3A4, CYP3A5, CYP3A7, and CYP3A43. CYP3A enzymes are indiscriminate toward substrates and are unique in that these enzymes metabolize both endogenous compounds and diverse xenobiotics. Constitutive regulation of CYP3A4 transcription, both positive and negative, is mediated by hepatocyte nuclear factor 4α (HNF4α) and other hepatic transcription factors. CYP3A4 expression is modulated by various mechanisms involving nuclear receptors, hormones, xenobiotics, and signaling molecules. CYP3A4 is regulated by a large number of xenobiotics, including many drugs, endogenous compounds, and many hormones, such as triiodothyronine, dexamethasone, and growth hormone. Xenobiotic- and endobiotic-mediated CYP3A4 induction is indirect and entails activation of such ligand-dependent nuclear receptors as PXR, CAR, VDR, glucocorticoid receptor (GR) α, estrogen receptor (ER) α, bile acid receptor (farnesoid X receptor; FXR), oxysterol receptor (liver X receptor; LXR), and peroxisome proliferator-activated receptor alpha (PPARα) as well as by binding to the three major cis-acting modules: CLEM4, distal XREM, and prPXRE.
1. Introduction
The CYP3A subfamily is affiliated with the cytochrome P450 (CYP) superfamily, which represents monooxygenases that catalyze the breakdown of various substances via hydroxylation and epoxidation with the participation of an electron donor (NADPH) and molecular oxygen [1]. CYP enzymes function as the first line of defense against exogenous chemical agents [2]. CYP enzymes are responsible for approximately three-quarters of all drug metabolism reactions in the human body [3][4]. CYP enzymes are involved in many critical metabolic reactions, including the metabolism of steroid hormones, bile acids, polyunsaturated fatty acids, leukotrienes, and eicosanoids [3].
Genes of CYP enzymes have been found in the genetic material of representatives of all kingdoms of living organisms, including plants. There are 57 known functional CYP genes in the human genome, aside from 58 pseudogenes whose protein products are enzymes metabolizing a wide range of endogenous and exogenous chemical compounds [2][5][6]. The genes of CYP enzymes are categorized into 18 families and 43 subfamilies based on the percentage of amino acid sequence homology. Just 3 families—CYP2, CYP3, and CYP4—contain more genes than the other 15 families combined [4][7]. The human CYP3 family consists of a single subfamily, CYP3A, which contains four genes (CYP3A4, CYP3A5, CYP3A7, and CYP3A43) encoding four functional enzymes [5][6][8][9][10].
CYP3A is a major subfamily in the cytochrome P450 superfamily. CYP3A enzymes are involved in the metabolism of more than 30% [11] and according to other reports 45–60% [12][13][14] of all pharmaceutical drugs currently on the market. CYP3A enzymes also metabolize some endogenous substrates, including hormones and bile acids, as well as nonpharmaceutical xenobiotics [11][12][13].
Expression of CYP3A enzymes is regulated and varies under the influence of various exogenous (drugs, chemicals, and diets) and endogenous factors (fatty acids, hormones, cytokines, and microRNAs [miRs or miRNAs]) [11].
CYP3A enzymes’ activity can be influenced by anthropogenic environmental chemicals: organophosphates, carbamates, parabens, benzotriazole UV stabilizers, and plasticizers [11][12]. Natural compounds present in foods—e.g., flavonoids found in fruits and vegetables, coffee, tea, chocolate, and wine—can alter CYP3A enzymes’ expression [15]. A prime example is the inhibition of CYP3A enzymes’ expression by components of grapefruit juice [12][16]. There is experimental evidence that retinoids can regulate the expression of CYP3A genes [17]. Certain diets, such as high-fat diets, can alter the expression of CYP3A genes [18], and it is likely that human dietary habits can affect basal expression of these genes [11].
Many of these substances are in turn metabolized by induced CYP3A enzymes, and this feedback mechanism implements detoxification of potentially harmful compounds [12].
2. Mechanisms of CYP3A Regulation
CYP3A4 expression is modulated by various mechanisms involving nuclear receptors, hormones, xenobiotics, and signaling molecules. CYP3A4 is regulated by a large number of xenobiotics, including many drugs, endogenous compounds, and many hormones, such as triiodothyronine, dexamethasone, and growth hormone [19].
Xenobiotic- and endobiotic-mediated CYP3A4 induction is indirect and entails activation of such ligand-dependent nuclear receptors as PXR, CAR, VDR, glucocorticoid receptor (GR) α, estrogen receptor (ER) α, bile acid receptor (farnesoid X receptor; FXR), oxysterol receptor (liver X receptor; LXR), and peroxisome proliferator-activated receptor alpha (PPARα) [10][11][14][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34] as well as by binding to the three major cis-acting modules: CLEM4, distal XREM, and prPXRE (Figure 1) [10].
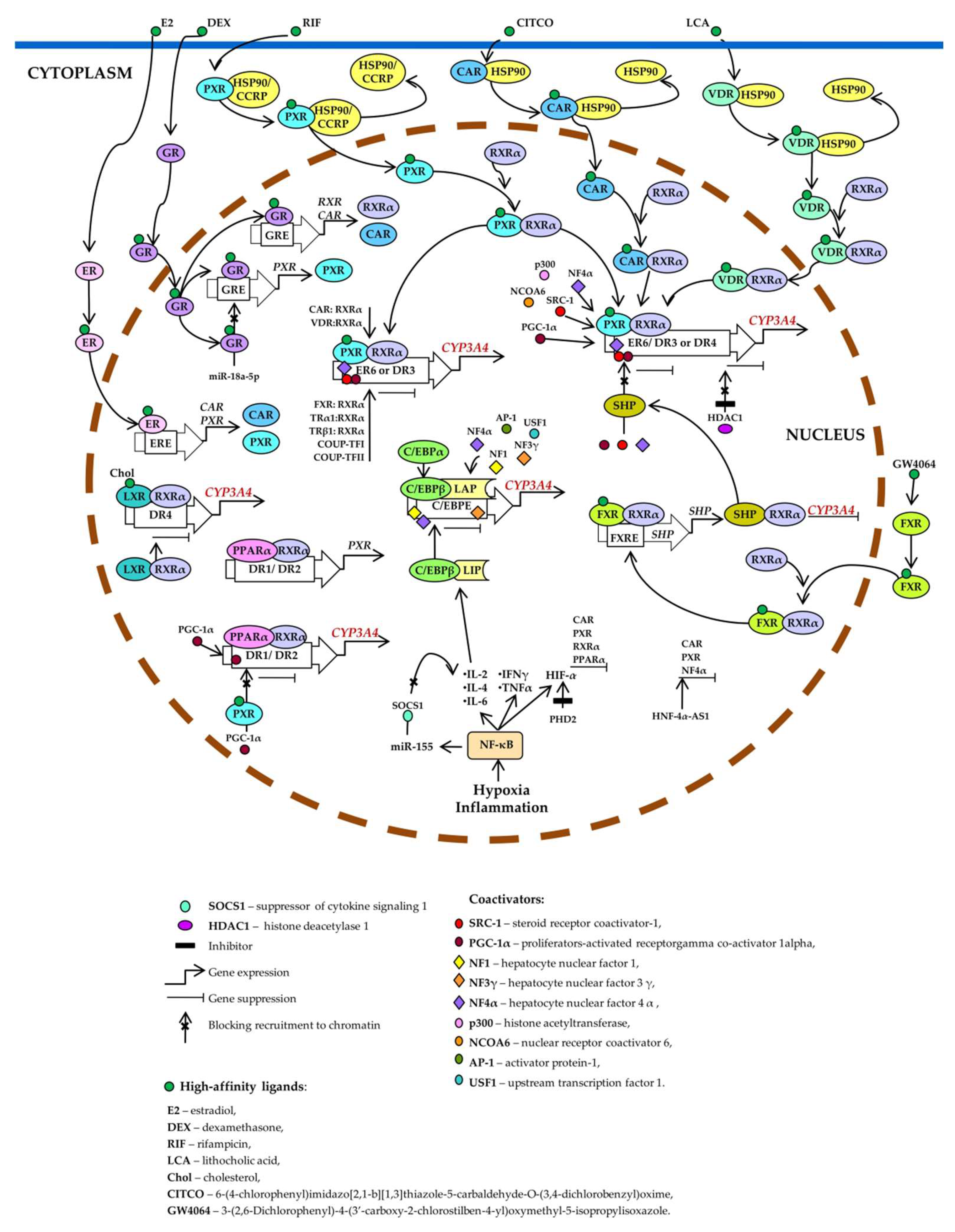
Figure 1. Transcriptional regulation of CYP3A4. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and vitamin D receptor (VDR) control basal and inducible expression of CYP3A4 through competitive binding to the same set of response elements (everted repeats 6, ER6; direct repeats DR3, and DR4). PXR, CAR, or VDR unbound by a ligand is located in the cytoplasm as a complex with heat shock protein 90 (HSP90) or cytoplasmic constitutive active/androstane receptor retention protein (CCRP). When activated by a ligand, each of them forms a heterodimer with retinoid X receptor α (RXRα), relocates to the nucleus, binds to a response element, recruits coactivators, and activates CYP3A4 transcription. Estrogen receptor (ER) and glucocorticoid receptor (GR) raise CYP3A4 expression by enhancing the expression of CAR, RXRα, and PXR. Ligand-activated farnesoid X receptor (FXR) upregulates small heterodimer partner (SHP), which prevents the recruitment of coactivators to chromatin and/or forms heterodimers with RXRα, thereby inhibiting CYP3A4 expression. Histone deacetylase 1 (HDAC1) inhibition by carbamazepine downregulates CYP3A4. Liver X receptor (LXR) forms a heterodimer with RXRα that then binds to DR4 in the target gene, thus repressing its expression. After the binding of a ligand to LXR or RXR, the heterodimer changes its conformation, which leads to a release of corepressors and the recruitment of coactivators. This event causes transcription of a target gene (peroxisome proliferator-activated receptor alpha; PPARα), its protein product binds as a PPARα–RXRα heterodimer to motifs DR1 and DR2 and enhances the transcription of CYP3A4 and PXR. Ligand-activated PXR suppresses PPARα-dependent gene expression by inhibiting peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α) recruitment. Hypoxia and inflammation induce the activity of nuclear factor kappa B (NF-κB) and promote a release of the cytokines that increase the transcription of CCAAT enhancer-binding protein beta (C/EBPβ) and the translation of C/EBPβ-LIP mRNA. C/EBPβ-LIP competes with C/EBPα and C/EBPβ-LAP for binding to response elements in the promoter of CYP3A4, thus lowering its expression. NF-κB activates miR-155, which directly targets mRNAs of suppressors of cytokine signaling proteins (especially suppressor of cytokine signaling 1: SOCS1) thereby inhibiting obligatory negative feedback regulation of inflammatory responses. Abbreviations. C/EBPβ-LAP: a C/EBPβ isoform called liver-enriched activator protein; C/EBPβ-LIP: a C/EBPβ isoform called liver-enriched inhibitory protein; COUP-TFI: chicken ovalbumin upstream promoter transcription factor I; COUP-TFII: chicken ovalbumin upstream promoter transcription factor II; DR1, DR2, DR3, DR4, and ER6: AG(G/T)TCA-like direct repeats separated by 1, 2, 3, or 4 bases, respectively, and an inverted repeat separated by 6 bases; ERE: ER-responsive element; FXRE: FXR-responsive element, GRE: GR-responsive element; HIF-1α: hypoxia-inducible factor 1-α; HNF-4α-AS1: hepatocyte nuclear factor 4α-antisense-RNA 1; IL-2, -4, or -6: interleukin 2, 4 or 6; INFγ: interferon γ; PHD2: prolyl hydroxylase domain-containing protein 2; TNF: tumor necrosis factor; TRα1: thyroid hormone receptor-α1; TRβ1: thyroid hormone receptor-β1.
Thus, most CYP3A inducers act through transcriptional activation [9][11][13]. CYP3A isoforms and nuclear receptors involved in their regulation are subject (as part of post-transcriptional regulation) to ubiquitination (CYP3A4 and CYP3A5) and phosphorylation (CYP3A4 and PXR) [11], whereas post-translational regulation of CYP3A enzymes consists of the stabilization of CYP3A mRNAs and proteins [6][11].
Molecular mechanisms of induction may differ among the four major human CYP3A genes and among their polymorphic variants owing to differences in their structure, and the mechanisms can also differ among different tissues, possibly because of different ratios of crucial protein factors. This complexity is a consequence of the wide range of CYP3A ligands and of nuclear receptors mediating the induction of CYP3A genes [9][13].
3. Involvement of CYP3A Enzymes in Biological Processes
CYP3A enzymes have very broad substrate specificity and metabolize a wide range of compounds in terms of chemical and biological properties. They catalyze reactions of hydroxylation, N-demethylation, O-dealkylation, S-oxidation, deamination, and epoxidation [35] of endogenous and exogenous compounds. CYP3A perform physiological functions by taking part in such endogenous processes as steroid catabolism, bile acid metabolism, and lipid and vitamin D metabolism. CYP3A enzymes metabolize a wide variety of therapeutics and may play an important role in alterations of biological activities of drugs or in enhanced clearance of drugs as well as in drug interactions. For instance, CYP3A enzymes’ substrates are such endogenous compounds as hormones, cholesterol, bile acids, arachidonic acid, and vitamin D as well as the vast majority of drugs and of xenobiotics that are not pharmaceuticals [11][12][13].
4. CYP3A Involvement in Pathological Processes
Cholestasis is a pathological condition where normal flow of bile is low or disturbed, and bile acids accumulate in the liver. Stimulation of CYP3A4 activity in cholestasis may be an effective therapeutic approach to such diseases [19].
Arachidonic acid metabolites known as eicosanoids (EETs), whose emergence depends on CYP3A-mediated metabolism, are implicated in the pathophysiology of various diseases. For example, the CYP3A4 epoxygenase, responsible for the production of EETs, is overexpressed in breast cancer and is linked with the initiation and progression of breast cancer [36]. In human hepatoma cell line Hep3B, overexpression of CYP3A4 also promotes cell growth and cell cycle transition from the G1 to S phase [37].
The role of CYP3A enzymes in the metabolism of sex steroid hormones implies an association with the development of hormone-dependent diseases. It has long been known that CYP3A enzymes are expressed in normal and tumorous tissues of the breast [38][39][40] and of the prostate [41][42], in cells of the endometrium and cervix [43][44], and in ovarian tumors [45].
Vitamin D has multiple effects on the biological processes that regulate the metabolism of calcium and phosphorus and also affects proliferation, differentiation, and apoptosis of cells as well as immune regulation [46][47][48][49][50][51][52][53]. CYP3A4, by taking part in the inactivation of an active vitamin D metabolite [1.25(ОН)2D3], may have a significant impact on circulating vitamin D levels [54][55].
It is now clear that the expression and activity of CYP3A enzymes are affected by such pathological conditions as infection, inflammation, and cancer [10][56][57].
5. Concluding Remarks
This research is aimed at highlighting the main roles of CYP3A enzymes along with their unique characteristics in the metabolism of biologically active endogenous compounds and numerous xenobiotics that are important in clinical pharmacology as well as the involvement of these enzymes in a wide range of physiological and pathological phenomena. The scientific literature cited in this research attests to remarkable efforts and advances in the understanding how the CYP3A family of phase I biotransformation enzymes is integrated into the vast and complex network of physiological processes detoxifying endo- and xenobiotics. The function of CYP3A enzymes is complex because the effects of activation their genes are determined by a wide range of endogenous and exogenous ligands and by a unique regulatory system that involves CYP3A enzymes in many physiological and pathological processes in cells and tissues of the body (Figure 2).

Figure 2. Effects of CYP3A enzymes on the metabolism of endo- and xenobiotics have an influence on a wide range of physiological and pathophysiological processes in the body.
The totality of evidence indicates that the activation of CYP3A genes can be either beneficial or detrimental during diseases of various organs and tissues. The ultimate effects depend both on the context of a disease and on the nature of ligands of the nuclear receptors that control CYP3A genes’ transcription.
Currently, the molecular mechanisms by which CYP3A enzymes take part in pathogenesis are well understood only for a few diseases; in particular, a role of CYP3A5 in carcinogenesis has been demonstrated. There are more reports of (i) diseases associated with the participation of CYP3A enzymes in the metabolism of endogenous compounds and (ii) pathological conditions affecting the expression and activity of CYP3A enzymes. The consequence of an alteration of these enzymes’ activities is a change in the pharmacokinetics of the drugs used for treatment. Much basic research has been conducted on the role of CYP3A enzymes in pathological processes, but clinical studies that are aimed at influencing the mechanisms of signaling pathways regulating CYP3A genes in various diseases are still insufficient, and further investigation is needed.
References
- F. Peter Guengerich; Mechanisms of Cytochrome P450-Catalyzed Oxidations. ACS Catalysis 2018, 8, 10964-10976, 10.1021/acscatal.8b03401.
- Andrew J. Annalora; Craig B. Marcus; Patrick L. Iversen; Alternative Splicing in the Cytochrome P450 Superfamily Expands Protein Diversity to Augment Gene Function and Redirect Human Drug Metabolism. Drug Metabolism and Disposition 2017, 45, 375-389, 10.1124/dmd.116.073254.
- Timothy S. Tracy; Amarjit S. Chaudhry; Bhagwat Prasad; Kenneth E. Thummel; Erin G. Schuetz; Xiao-Bo Zhong; Yun-Chen Tien; Hyunyoung Jeong; Xian Pan; Laura M. Shireman; et al.Jessica Tay-SontheimerYvonne S. Lin Interindividual Variability in Cytochrome P450-Mediated Drug Metabolism. Drug Metabolism and Disposition 2015, 44, 343-351, 10.1124/dmd.115.067900.
- Rosemary H. Waring; Cytochrome P450: genotype to phenotype. Xenobiotica 2019, 50, 9-18, 10.1080/00498254.2019.1648911.
- Xiaoping Chen; Haijian Wang; Gangqiao Zhou; Xiumei Zhang; Xiaojia Dong; Lianteng Zhi; Li Jin; Fuchu He; Molecular Population Genetics of HumanCYP3ALocus: Signatures of Positive Selection and Implications for Evolutionary Environmental Medicine. Environmental health perspectives 2009, 117, 1541-1548, 10.1289/ehp.0800528.
- G. G. Gibson; N. J. Plant; K. E. Swales; A. Ayrton; W. El-Sankary; Receptor-dependent transcriptional activation of cytochrome P4503A genes: induction mechanisms, species differences and interindividual variation in man. Xenobiotica 2002, 32, 165-206, 10.1080/00498250110102674.
- Daniel W. Nebert; Kjell Wikvall; Walter L. Miller; Human cytochromes P450 in health and disease. Philosophical Transactions B 2013, 368, 20120431, 10.1098/rstb.2012.0431.
- Ann K. Daly; Ann K Daly; Significance of the Minor Cytochrome P450 3A Isoforms. Clinical Pharmacokinetics 2006, 45, 13-31, 10.2165/00003088-200645010-00002.
- Hannu Raunio; Jukka Hakkola; Olavi Pelkonen; Regulation of CYP3A genes in the human respiratory tract. Chemico-Biological Interactions 2005, 151, 53-62, 10.1016/j.cbi.2003.12.007.
- Ulrich M. Zanger; Matthias Schwab; Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacology & Therapeutics 2013, 138, 103-141, 10.1016/j.pharmthera.2012.12.007.
- Chieri Fujino; Seigo Sanoh; Toshiya Katsura; Variation in Expression of Cytochrome P450 3A Isoforms and Toxicological Effects: Endo- and Exogenous Substances as Regulatory Factors and Substrates. Biological & pharmaceutical bulletin 2021, 44, 1617-1634, 10.1248/bpb.b21-00332.
- Oliver Burk; Leszek Wojnowski; Cytochrome P450 3A and their regulation. Naunyn-Schmiedebergs Archives of Pharmacology 2004, 369, 105-124, 10.1007/s00210-003-0815-3.
- Xuan Qin; Xin Wang; Role of vitamin D receptor in the regulation of CYP3A gene expression. Acta Pharmaceutica Sinica. B 2019, 9, 1087-1098, 10.1016/j.apsb.2019.03.005.
- Scott R. Penzak; Carlos Rojas‐Fernandez; 4β‐Hydroxycholesterol as an Endogenous Biomarker for CYP3A Activity: Literature Review and Critical Evaluation. The Journal of Clinical Pharmacology 2018, 59, 611-624, 10.1002/jcph.1391.
- Masayuki Tsujimoto; Maya Horie; Hiroko Honda; Kohji Takara; Kohshi Nishiguchi; The Structure-Activity Correlation on the Inhibitory Effects of Flavonoids on Cytochrome P450 3A Activity. Biological & pharmaceutical bulletin 2009, 32, 671-676, 10.1248/bpb.32.671.
- Ken-Ichi Fujita; FOOD-DRUG INTERACTIONS VIA HUMAN CYTOCHROME P450 3A (CYP3A). Drug metabolism and drug interactions 2004, 20, 195-218, 10.1515/dmdi.2004.20.4.195.
- Shiyong Chen; Kun Wang; Yu-Jui Yvonne Wan; Retinoids activate RXR/CAR-mediated pathway and induce CYP3A. Biochemical pharmacology 2010, 79, 270-276, 10.1016/j.bcp.2009.08.012.
- Xilin Li; Zemin Wang; James E. Klaunig; Modulation of xenobiotic nuclear receptors in high-fat diet induced non-alcoholic fatty liver disease. Toxicology 2018, 410, 199-213, 10.1016/j.tox.2018.08.007.
- Chen Jiezhong; Zhao Kong-Nan; Jiezhong Chen; The role of CYP3A4 in the biotransformation of bile acids and therapeutic implication for cholestasis.. Annals of Translational Medicine 2014, 2, 7-7, 10.3978/j.issn.2305-5839.2013.03.02.
- L C Quattrochi; P S Guzelian; Cyp3A regulation: from pharmacology to nuclear receptors.. Drug Metabolism and Disposition 2001, 29, 615-22.
- Brahim Achour; Jill Barber; Amin Rostami-Hodjegan; Expression of Hepatic Drug-Metabolizing Cytochrome P450 Enzymes and Their Intercorrelations: A Meta-Analysis. Drug Metabolism and Disposition 2014, 42, 1349-1356, 10.1124/dmd.114.058834.
- Bryan Goodwin; Ecushla Hodgson; Christopher Liddle; The Orphan Human Pregnane X Receptor Mediates the Transcriptional Activation ofCYP3A4by Rifampicin through a Distal Enhancer Module. Molecular Pharmacology 1999, 56, 1329-1339, 10.1124/mol.56.6.1329.
- Ramiro Jover; Roque Bort; Maria J. Gomez‐Lechon; Jose V. Castell; Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: A study using adenovirus-mediated antisense targeting. Hepatology 2001, 33, 668-675, 10.1053/jhep.2001.22176.
- Maria Thomas; Oliver Burk; Britta Klumpp; Benjamin A. Kandel; Georg Damm; Thomas S. Weiss; Kathrin Klein; Matthias Schwab; Ulrich M. Zanger; Direct Transcriptional Regulation of Human Hepatic Cytochrome P450 3A4 (CYP3A4) by Peroxisome Proliferator–Activated Receptor Alpha (PPARα). Molecular Pharmacology 2013, 83, 709-718, 10.1124/mol.112.082503.
- Jean-Marc Pascussi; Lionel Drocourt; Sabine Gerbal-Chaloin; Jean-Michel Fabre; Patrick Maurel; Marie-José Vilarem; Dual effect of dexamethasone onCYP3A4gene expression in human hepatocytes. Journal of Biological Inorganic Chemistry 2001, 268, 6346-6358, 10.1046/j.0014-2956.2001.02540.x.
- Zofia Duniec-Dmuchowski; Ewa Ellis; Stephen C. Strom; Thomas A. Kocarek; Regulation of CYP3A4 and CYP2B6 expression by liver X receptor agonists. Biochemical pharmacology 2007, 74, 1535-1540, 10.1016/j.bcp.2007.07.040.
- Danxin Wang; Rong Lu; Grzegorz Rempala; Wolfgang Sadee; Ligand-Free Estrogen Receptor α (ESR1) as Master Regulator for the Expression of CYP3A4 and Other Cytochrome P450 Enzymes in the Human Liver. Molecular Pharmacology 2019, 96, 430-440, 10.1124/mol.119.116897.
- Alejandro Carazo; Lucie Hyrsova; Jan Dusek; Hana Chodounska; Alzbeta Horvatova; Karel Berka; Vaclav Bazgier; Hongying Gan-Schreier; Waleé Chamulitrat; Eva Kudova; et al.Petr Pavek Acetylated deoxycholic (DCA) and cholic (CA) acids are potent ligands of pregnane X (PXR) receptor. Toxicology letters 2017, 265, 86-96, 10.1016/j.toxlet.2016.11.013.
- Carmela Gnerre; Sharon Blättler; Michel R Kaufmann; Renate Looser; Urs A Meyer; Regulation of CYP3A4 by the bile acid receptor FXR. Pharmacogenetics 2004, 14, 635-645, 10.1097/00008571-200410000-00001.
- Ryutaro Adachi; Yoshio Honma; Hiroyuki Masuno; Katsuyoshi Kawana; Iichiro Shimomura; Sachiko Yamada; Makoto Makishima; Selective activation of vitamin D receptor by lithocholic acid acetate, a bile acid derivative. Journal of lipid research 2005, 46, 46-57, 10.1194/jlr.m400294-jlr200.
- Jean-Marc Pascussi; Sabine Gerbal-Chaloin; Cédric Duret; Martine Daujat-Chavanieu; Marie-José Vilarem; Patrick Maurel; The Tangle of Nuclear Receptors that Controls Xenobiotic Metabolism and Transport: Crosstalk and Consequences. Annual Review of Pharmacology and Toxicology 2008, 48, 1-32, 10.1146/annurev.pharmtox.47.120505.105349.
- Tsutomu Matsubara; Kouichi Yoshinari; Kazunobu Aoyama; Mika Sugawara; Yuji Sekiya; Kiyoshi Nagata; Yasushi Yamazoe; Role of Vitamin D Receptor in the Lithocholic Acid-Mediated CYP3A Induction in Vitro and in Vivo. Drug Metabolism and Disposition 2008, 36, 2058-2063, 10.1124/dmd.108.021501.
- Adrian Schröder; Johannes Wollnik; Clemens Wrzodek; Andreas Dräger; Michael Bonin; Oliver Burk; Maria Thomas; Wolfgang E. Thasler; Ulrich M. Zanger; Andreas Zell; et al. Inferring statin-induced gene regulatory relationships in primary human hepatocytes. Bioinformatics 2011, 27, 2473-2477, 10.1093/bioinformatics/btr416.
- Maryam Rakhshandehroo; Guido Hooiveld; Michael Müller; Sander Kersten; Comparative Analysis of Gene Regulation by the Transcription Factor PPARα between Mouse and Human. PLoS ONE 2009, 4, e6796-e6796, 10.1371/journal.pone.0006796.
- Ibrahim Ince; Catherijne A. J. Knibbe; Meindert Danhof; Saskia N. de Wildt; Developmental Changes in the Expression and Function of Cytochrome P450 3A Isoforms: Evidence from In Vitro and In Vivo Investigations. Clinical Pharmacokinetics 2013, 52, 333-345, 10.1007/s40262-013-0041-1.
- Esau Floriano-Sanchez; Noemi Cardenas Rodriguez; Cindy Bandala; Elvia Coballase-Urrutia; Jaime Lopez-Cruz; CYP3A4 Expression in Breast Cancer and its Association with Risk Factors in Mexican Women. Asian Pacific Journal of Cancer Prevention 2014, 15, 3805-3809, 10.7314/apjcp.2014.15.8.3805.
- Ami Oguro; Koichi Sakamoto; Yoshihiko Funae; Susumu Imaoka; Overexpression of CYP3A4, but not of CYP2D6, Promotes Hypoxic Response and Cell Growth of Hep3B Cells. Drug metabolism and pharmacokinetics 2011, 26, 407-415, 10.2133/dmpk.dmpk-11-rg-017.
- Heike Hellmold; Tove Rylander; Malin Magnusson; Eva Reihnér; Margaret Warner; Jan-Åke Gustafsson; Characterization of Cytochrome P450 Enzymes in Human Breast Tissue from Reduction Mammaplasties1. Journal of Clinical Endocrinology & Metabolism 1998, 83, 886-895, 10.1210/jcem.83.3.4647.
- Graeme I. Murray; Richard J. Weaver; Pamela J. Paterson; Stanley W. B. Ewen; William T. Melvin; M. Burke Danny; Expression of xenobiotic metabolizing enzymes in breast cancer. The Journal of Pathology 1993, 169, 347-353, 10.1002/path.1711690312.
- Yasuo Miyoshi; Akiko Ando; Yuuki Takamura; Tetsuya Taguchi; Yasuhiro Tamaki; Shinzaburo Noguchi; Prediction of response to docetaxel by CYP3A4 mRNA expression in breast cancer tissues. International Journal of Cancer 2001, 97, 129-132, 10.1002/ijc.1568.
- N. Finnström; C. Bjelfman; T. G. Söderström; Gillian Smith; Lars Egevad; B. J. Norlén; Charles Roland Wolf; A. Rane; Detection of cytochrome P450 mRNA transcripts in prostate samples by RT-PCR. European Journal of Clinical Investigation 2001, 31, 880-886, 10.1046/j.1365-2362.2001.00893.x.
- Graeme I. Murray; Valerie E. Taylor; Judith A. McKay; Richard J. Weaver; Stanley W. B. Ewen; William T. Melvin; M. Danny Burke; The immunohistochemical localization of drug-metabolizing enzymes in prostate cancer. The Journal of Pathology 1995, 177, 147-152, 10.1002/path.1711770208.
- Janne Hukkanen; Marjatta Mantyla; Lauri Kangas; Pauli Wirta; Jukka Hakkola; Pauliina Paakki; Sari Evisalmi; Olavi Pelkonen; Hannu Raunio; Expression of cytochrome P450 genes encoding enzymes active in the metabolism of tamoxifen in human uterine endometrium.. Basic & Clinical Pharmacology & Toxicology 1998, 82, 93-97, 10.1111/j.1600-0773.1998.tb01404.x.
- Mohamadi A. Sarkar; Vijay Vadlamuri; Shobha Ghosh; Douglas D. Glover; Expression and cyclic variability of CYP3A4 and CYP3A7 isoforms in human endometrium and cervix during the menstrual cycle.. Drug Metabolism and Disposition 2003, 31, 1-6, 10.1124/dmd.31.1.1.
- Julie A. DeLoia; William C. Zamboni; Jacqueline M. Jones; Sandra Strychor; Joseph L. Kelley; Holly H. Gallion; Expression and activity of taxane-metobolizing enzymes in ovarian tumors. Gynecologic Oncology 2008, 108, 355-360, 10.1016/j.ygyno.2007.10.029.
- Hanmin Wang; Weiwen Chen; Dongqing Li; Xiaoe Yin; Xiaode Zhang; Nancy Olsen; Song Guo Zheng; Vitamin D and Chronic Diseases. Aging and Disease 2017, 8, 346-353, 10.14336/ad.2016.1021.
- Michael F Holick; Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. The American journal of clinical nutrition 2004, 80, 1678S-1688S, 10.1093/ajcn/80.6.1678s.
- Chantal Mathieu; Klaus Badenhoop; Vitamin D and type 1 diabetes mellitus: state of the art. Trends in Endocrinology & Metabolism 2005, 16, 261-266, 10.1016/j.tem.2005.06.004.
- Nora Nikolac Gabaj; Adriana Unic; Marijana Miler; Tomislav Pavicic; Jelena Culej; Ivan Bolanca; Davorka Herman Mahecic; Lara Milevoj Kopcinovic; Alen Vrtaric; In sickness and in health: pivotal role of vitamin D. Biochemia medica 2020, 30, 202-214, 10.11613/bm.2020.020501.
- Sang-Min Jeon; Eun-Ae Shin; Exploring vitamin D metabolism and function in cancer. Experimental & Molecular Medicine 2018, 50, 1-14, 10.1038/s12276-018-0038-9.
- Alison M Mondul; Stephanie J Weinstein; Tracy M Layne; Demetrius Albanes; Vitamin D and Cancer Risk and Mortality: State of the Science, Gaps, and Challenges. Epidemiologic reviews 2017, 39, 28-48, 10.1093/epirev/mxx005.
- Karin Amrein; Mario Scherkl; Magdalena Hoffmann; Stefan Neuwersch-Sommeregger; Markus Köstenberger; Adelina Tmava Berisha; Gennaro Martucci; Stefan Pilz; Oliver Malle; Vitamin D deficiency 2.0: an update on the current status worldwide. European Journal of Clinical Nutrition 2020, 74, 1498-1513, 10.1038/s41430-020-0558-y.
- Sihe Wang; Epidemiology of vitamin D in health and disease. Nutrition research reviews 2009, 22, 188-203, 10.1017/s0954422409990151.
- Natasha Khazai; Suzanne E. Judd; Vin Tangpricha; Calcium and vitamin D: Skeletal and extraskeletal health. Current Rheumatology Reports 2008, 10, 110-117, 10.1007/s11926-008-0020-y.
- James C. Fleet; The role of vitamin D in the endocrinology controlling calcium homeostasis. Molecular and cellular endocrinology 2017, 453, 36-45, 10.1016/j.mce.2017.04.008.
- E. T. Morgan; Impact of Infectious and Inflammatory Disease on Cytochrome P450–Mediated Drug Metabolism and Pharmacokinetics. Clinical Pharmacology & Therapeutics 2009, 85, 434-438, 10.1038/clpt.2008.302.
- Morgan, E.T.. Drug Metabolism in Diseases; Xie, W., Eds.; Academic Press: Boston, 2017; pp. 21-58.
- Morgan, E.T.. Drug Metabolism in Diseases; Xie, W., Eds.; Academic Press: Boston, 2017; pp. 21-58.




