
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jong-Joo Cheong | -- | 4363 | 2022-11-10 09:43:09 | | | |
| 2 | Vivi Li | Meta information modification | 4363 | 2022-11-11 02:03:07 | | |
Video Upload Options
Plants respond to drought stress by producing abscisic acid, a chemical messenger that regulates gene expression and thereby expedites various physiological and cellular processes including the stomatal operation to mitigate stress and promote tolerance. To trigger or suppress gene transcription under drought stress conditions, the surrounding chromatin architecture must be converted between a repressive and active state by epigenetic remodeling, which is achieved by the dynamic interplay among DNA methylation, histone modifications, loop formation, and non-coding RNA generation. Plants can memorize chromatin status under drought conditions to enable them to deal with recurrent stress. Furthermore, drought tolerance acquired during plant growth can be transmitted to the next generation. The epigenetically modified chromatin architectures of memory genes under stressful conditions can be transmitted to newly developed cells by mitotic cell division, and to germline cells of offspring by overcoming the restraints on meiosis.
1. Introduction
2. Transcription of Drought-Responsive Genes
2.1. Drought-Responsive Genes
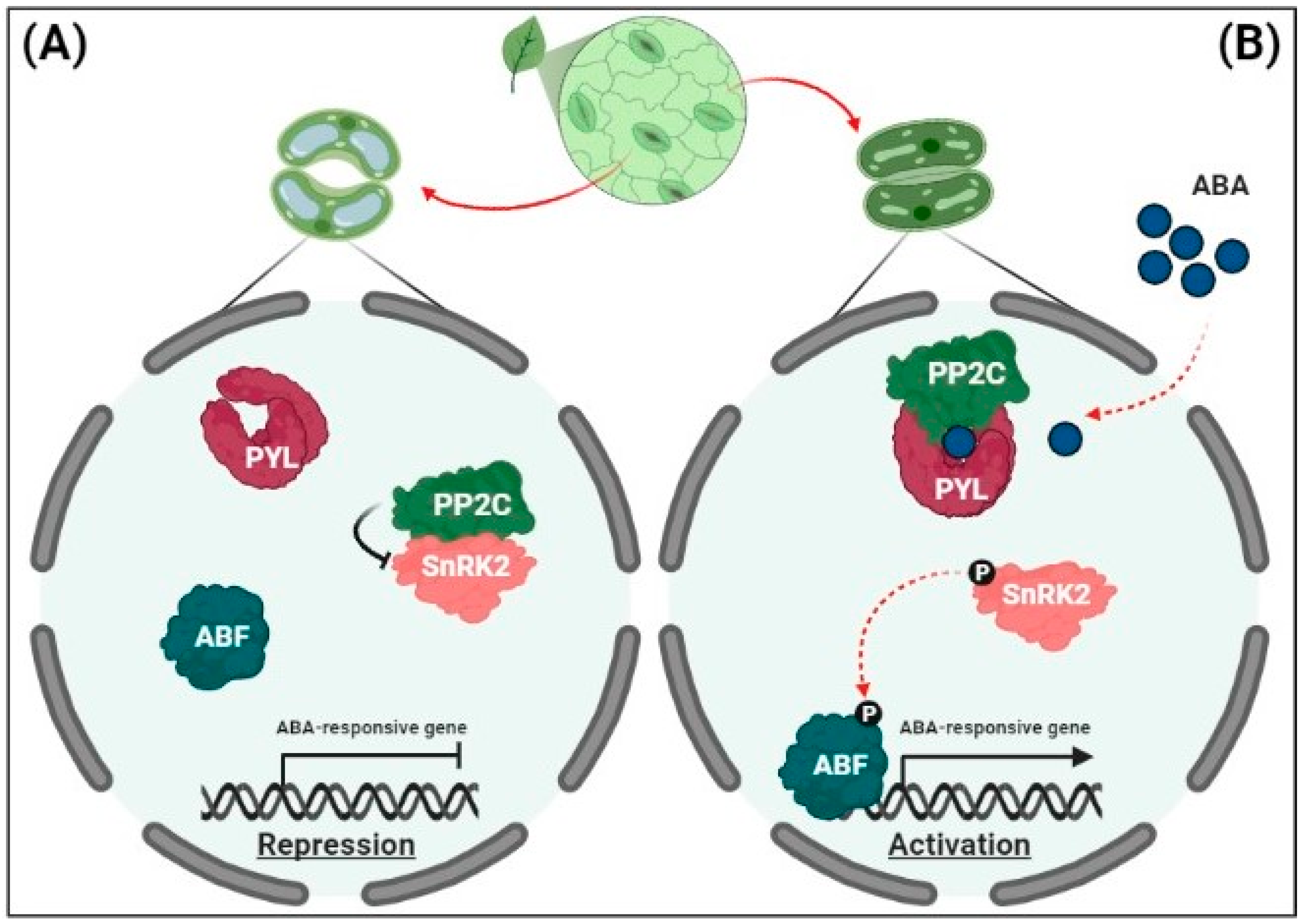
2.2. Regulation of ABA Signaling
3. Transcriptional Memory of Drought Tolerance
3.1. Drought Stress Memory
3.2. Drought Stress Memory Genes
3.3. Mechanism of Transcriptional Stress Memory
3.3.1. Epigenetic Marks for Stress Memory
4. Transgenerational Inheritance of Memory
4.1. Transgenerational Transmission of Drought Tolerance
4.2. DNA Methylation for Transgenerational Inheritance
4.3. Overcoming Meiosis
References
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 1996, 7, 161–167.
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Attal, N.B.; Venkateswarlu, B. Drought stress responses in crops. Funct. Integr. Genom. 2014, 14, 11–22.
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269.
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185.
- Bharath, P.; Gahir, S.; Raghavendra, A.S. Abscisic acid-induced stomatal closure: An important component of plant defense against abiotic and biotic stress. Front. Plant Sci. 2021, 12, 615114.
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genom. 2002, 2, 282–291.
- Hoth, S.; Morgante, M.; Sanchez, J.-P.; Hanafey, M.K.; Tingey, S.V.; Chua, N.H. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 2002, 115, 4891–4900.
- Cairns, B.R. The logic of chromatin architecture and remodeling at promoters. Nature 2009, 461, 193–198.
- Yamamuro, C.; Zhu, J.-K.; Yang, Z. Epigenetic modifications and plant hormone action. Mol. Plant 2016, 9, 57–70.
- Kim, J.-H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013.
- Ma, X.; Zhao, F.; Zhou, B. The characters of non-coding RNAs and their biological roles in plant development and abiotic stress response. Int. J. Mol. Sci. 2022, 23, 4124.
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608.
- Godwin, J.; Farrona, S. Plant epigenetic stress memory induced by drought: A physiological and molecular perspective. Methods Mol. Biol. 2020, 2093, 243–259.
- Choudhary, M.; Singh, A.; Rakshit, S. Coping with low moisture stress: Remembering and responding. Physiol. Plant. 2021, 172, 1162–1169.
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531.
- Kinoshita, T.; Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863.
- Marthandan, V.; Geetha, R.; Kumutha, K.; Renganathan, V.G.; Karthikeyan, A.; Ramalingam, J. Seed priming: A feasible strategy to enhance drought tolerance in crop plants. Int. J. Mol. Sci. 2020, 21, 8258.
- Luo, M.; Liu, X.; Singh, P.; Cui, Y.; Zimmerli, L.; Wu, K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 129–136.
- Kim, J.-M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 8, 114.
- Avramova, Z. Transcriptional ‘memory’ of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159.
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124.
- Crisp, P.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340.
- Sharma, M.; Kumar, P.; Verma, V.; Sharma, R.; Bhargava, B.; Irfan, M. Understanding plant stress memory response for abiotic stress resilience: Molecular insights and prospects. Plant Physiol. Biochem. 2022, 179, 10–24.
- Naumova, N.; Imakaev, M.; Fudenberg, G.; Zhan, Y.; Lajoie, B.R.; Mirny, L.A.; Dekker, J. Organization of the mitotic chromosome. Science 2013, 342, 948–953.
- Wang, F.; Higgins, J.M. Histone modifications and mitosis: Countermarks, landmarks, and bookmarks. Trends Cell Biol. 2013, 23, 175–184.
- Molinier, J.; Ries, G.; Zipfel, C.; Hohn, B. Transgeneration memory of stress in plants. Nature 2006, 442, 1046–1049.
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016, 50, 467–491.
- Springer, N.M. Epigenetics and crop improvement. Trends Genet. 2013, 29, 241–247.
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251.
- Bilichak, A.; Kovalchuk, I. Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 2016, 67, 2081–2092.
- Todaka, D.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. ABA-responsive gene expression in response to drought stress: Cellular regulation and long-distance signaling. Adv. Bot. Res. 2019, 92, 83–113.
- Cheong, J.-J. Modulation of abscisic acid signaling for stomatal operation under salt stress conditions. Adv. Bot. Res. 2022, 103, 89–121.
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94.
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730.
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl Acad. Sci. USA 2000, 97, 11632–11637.
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227.
- Cutler, A.; Krochko, J. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478.
- Merilo, E.; Jalakas, P.; Laanemets, K.; Mohammadi, O.; Hõrak, H.; Kollist, H.; Brosché, M. Abscisic acid transport and homeostasis in the context of stomatal regulation. Mol. Plant 2015, 8, 1321–1333.
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA transport and plant water stress responses. Trends Plant Sci. 2018, 23, 513–522.
- Seo, M.; Marion-Poll, A. Abscisic acid metabolism and transport. Adv. Bot. Res. 2019, 92, 1–49.
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496.
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.S.; Kollist, H.; Merilo, E.; et al. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 2019, 5, 1002–1011.
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593.
- Jung, C.; Nguyen, N.H.; Cheong, J.-J. Transcriptional regulation of protein phosphatase 2C genes to modulate abscisic acid signaling. Int. J. Mol. Sci. 2020, 21, 9517.
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839.
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signaling. Essays Biochem. 2015, 58, 29–48.
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49.
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685.
- Liu, H.; Able, A.J.; Able, J.A. Priming crops for the future: Rewiring stress memory. Trends Plant Sci. 2022, 27, 699–716.
- Sadhukhan, A.; Prasad, S.S.; Mitra, J.; Siddiqui, N.; Sahoo, L.; Kobayashi, Y.; Koyama, H. How do plants remember drought? Planta 2022, 256, 7.
- Walter, J.; Nagy, L.; Hein, R.; Rascher, U.; Beierkuhnlein, C.; Willner, E.; Jentsch, A. Do plants remember drought? Hints towards a drought-memory in grasses. Environ. Exp. Bot. 2011, 71, 34–40.
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740.
- Wang, X.; Vignjevic, M.; Jiang, D.; Jacobsen, S.; Wollenweber, B. Improved tolerance to drought stress after anthesis due to priming before anthesis in wheat (Triticum aestivum L.) var. Vinjett. J. Exp. Bot. 2014, 65, 6441–6456.
- Ramírez, D.A.; Rolando, J.L.; Yactayo, W.; Monneveux, P.; Mares, V.; Quiroz, R. Improving potato drought tolerance through the induction of long-term water stress memory. Plant Sci. 2015, 238, 26–32.
- Abdallah, M.B.; Methenni, K.; Nouairi, I.; Zarrouk, M.; Youssef, N.B. Drought priming improves subsequent more severe drought in a drought-sensitive cultivar of olive cv. Chétoui. Sci. Hortic. 2017, 221, 43–52.
- Tabassum, T.; Farooq, M.; Ahmad, R.; Zohaib, A.; Wahid, A.; Shahid, M. Terminal drought and seed priming improves drought tolerance in wheat. Physiol. Mol. Biol. Plants 2018, 24, 845–856.
- Goh, C.-H.; Nam, H.G.; Park, Y.S. Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 2003, 36, 240–255.
- Virlouvet, L.; Fromm, M. Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol. 2015, 205, 596–607.
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 2019, 10, 55.
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229.
- Kim, Y.-K.; Chae, S.; Oh, N.-I.; Nguyen, N.H.; Cheong, J.-J. Recurrent drought conditions enhance the induction of drought stress memory genes in Glycine max L. Front. Genet. 2020, 11, 576086.
- Ding, Y.; Virlouvet, L.; Liu, N.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 141.
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome response to drought, rehydration and re-dehydration in potato. Int. J. Mol. Sci. 2020, 21, 159.
- Brzezinka, K.; Altmann, S.; Czesnick, H.; Nicolas, P.; Gorka, M.; Benke, E.; Kabelitz, T.; Jähne, F.; Graf, A.; Kappel, C.; et al. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 2016, 5, e17061.
- Liu, J.; He, Z. Small DNA methylation, big player in plant abiotic stress responses and memory. Front. Plant Sci. 2020, 11, 595603.
- Wang, W.-S.; Pan, Y.-J.; Zhao, X.-Q.; Dwivedi, D.; Zhu, L.-H.; Ali, J.; Fu, B.-Y.; Li, Z.-K. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 1951–1960.
- Kou, S.Y.; Gu, Q.Y.; Duan, L.; Liu, G.J.; Yuan, P.R.; Li, H.H.; Wu, Z.G.; Liu, W.H.; Huang, P.; Liu, L. Genome-wide bisulphite sequencing uncovered the contribution of DNA methylation to rice short-term drought memory formation. J. Plant Growth Regul. 2021, 1–15.
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423.
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349.
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518.
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356.
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678.
- Blackledge, N.P.; Klose, R.J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833.
- Baile, F.; Gómez-Zambrano, Á.; Calonje, M. Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants. Plant Commun. 2022, 3, 100267.
- Bratzel, F.; López-Torrejón, G.; Koch, M.; Del Pozo, J.C.; Calonje, M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 2010, 20, 1853–1859.
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329.
- Wang, Q.; Shen, W.-H. Chromatin modulation and gene regulation in plants: Insight about PRC1 function. Biochem. Soc. Trans. 2018, 46, 957–966.
- Vijayanathan, M.; Trejo-Arellano, M.G.; Mozgová, I. Polycomb repressive complex 2 in eukaryotes-An evolutionary perspective. Epigenomes 2022, 6, 3.
- Mozgova, I.; Hennig, L. The Polycomb group protein regulatory network. Annu. Rev. Plant Biol. 2015, 66, 269–296.
- Bieluszewski, T.; Xiao, J.; Yang, Y.; Wagner, D. PRC2 activity, recruitment, and silencing: A comparative perspective. Trends Plant Sci. 2021, 26, 1186–1198.
- Godwin, J.; Farrona, S. The importance of networking: Plant Polycomb Repressive Complex 2 and its interactors. Epigenomes 2022, 6, 8.
- Mylne, J.S.; Barrett, L.; Tessadori, F.; Mesnage, S.; Johnson, L.; Bernatavichute, Y.V.; Jacobsen, S.E.; Fransz, P.; Dean, C. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl. Acad. Sci. USA 2006, 103, 5012–5017.
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-binding factor basic pentacysteine6 recruits the polycomb-repressive complex1 component like heterochromatin protein1 to GAGA DNA motifs. Plant Physiol. 2015, 168, 1013–1024.
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; Citerne, S.; Bendahmane, A.; Hirt, H.; et al. The Polycomb protein LHP1 regulates Arabidopsis thaliana stress responses through the repression of the MYC2-dependent branch of immunity. Plant J. 2019, 100, 1118–1131.
- Liu, N.; Ding, Y.; Fromm, M.; Avramova, Z. Different gene-specific mechanisms determine the ‘revised-response’ memory transcription patterns of a subset of A. thaliana dehydration stress responding genes. Nucleic Acids Res. 2014, 42, 5556–5566.
- Liu, N.; Fromm, M.; Avramova, Z. H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol. Plant 2014, 7, 502–513.
- Kim, J.-M.; To, T.K.; Ishida, J.; Matsui, A.; Kimura, H.; Seki, M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 847–856.
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016, 352, aad9780.
- Schuettengruber, B.; Bourbon, H.-M.; Croce, L.D.; Cavalli, G. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 2017, 171, 34–57.
- Fletcher, J.C. State of the art: TrxG factor regulation of post-embryonic plant development. Front. Plant Sci. 2017, 8, 1925.
- Grini, P.E.; Thorstensen, T.; Alm, V.; Vizcay-Barrena, G.; Windju, S.S.; Jørstad, T.S.; Wilson, Z.A.; Aalen, R.B. The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS ONE 2009, 4, e7817.
- Guo, L.; Yu, Y.; Law, J.A.; Zhang, X. SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 18557–18562.
- Carter, B.; Henderson, J.T.; Svedin, E.; Fiers, M.; McCarthy, K.; Smith, A.; Guo, C.; Bishop, B.; Zhang, H.; Riksen, T.; et al. Cross-talk between sporophyte and gametophyte generations is promoted by CHD3 chromatin remodelers in Arabidopsis thaliana. Genetics 2016, 203, 817–829.
- Chen, L.-Q.; Luo, J.-H.; Cui, Z.-H.; Xue, M.; Wang, L.; Zhang, X.-Y.; Pawlowski, W.P.; He, Y. ATX3, ATX4, and ATX5 encode putative H3K4 methyltransferases and are critical for plant development. Plant Physiol. 2017, 174, 1795–1806.
- Ding, Y.; Ndamukong, I.; Xu, Z.; Lapko, H.; Fromm, M.; Avramova, Z. ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 2012, 8, e1003111.
- Fromm, M.; Avramova, Z. ATX1/AtCOMPASS and the H3K4me3 marks: How do they activate Arabidopsis genes? Curr. Opin. Plant Biol. 2014, 21, 75–82.
- Howe, F.S.; Fischl, H.; Murray, S.C.; Mellor, J. Is H3K4me3 instructive for transcription activation? Bioessays 2017, 39, 1–12.
- Boyko, A.; Kovalchuk, I. Genome instability and epigenetic modification--heritable responses to environmental stress? Curr. Opin. Plant Biol. 2011, 14, 260–266.
- Verkest, A.; Byzova, M.; Martens, C.; Willems, P.; Verwulgen, T.; Slabbinck, B.; Rombaut, D.; Van de Velde, J.; Vandepoele, K.; Standaert, E.; et al. Selection for improved energy use efficiency and drought tolerance in canola results in distinct transcriptome and epigenome changes. Plant Physiol. 2015, 168, 1338–1350.
- Tabassum, T.; Farooq, M.; Ahmad, R.; Zohaib, A.; Wahid, A. Seed priming and transgenerational drought memory improves tolerance against salt stress in bread wheat. Plant Physiol. Biochem. 2017, 118, 362–369.
- Raju, S.K.K.; Shao, M.R.; Sanchez, R.; Xu, Y.Z.; Sandhu, A.; Graef, G.; Mackenzie, S. An epigenetic breeding system in soybean for increased yield and stability. Plant Biotechnol. J. 2018, 16, 1836–1847.
- Campos, E.I.; Stafford, J.M.; Reinberg, D. Epigenetic inheritance: Histone bookmarks across generations. Trends Cell Biol. 2014, 24, 664–674.
- Fabrizio, P.; Garvis, S.; Palladino, F. Histone methylation and memory of environmental stress. Cells 2019, 8, 339.
- Šrut, M. Ecotoxicological epigenetics in invertebrates: Emerging tool for the evaluation of present and past pollution burden. Chemosphere 2021, 282, 131026.
- Zenk, F.; Loeser, E.; Schiavo, R.; Kilpert, F.; Bogdanović, O.; Iovino, N. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 2017, 357, 212–216.
- Molla-Herman, A.; Matias, N.R.; Huynh, J.R. Chromatin modifications regulate germ cell development and transgenerational information relay. Curr. Opin. Insect Sci. 2014, 1, 10–18.
- Weiser, N.E.; Kim, J.K. Multigenerational regulation of the Caenorhabditis elegans chromatin landscape by germline small RNAs. Annu. Rev. Genet. 2019, 53, 289–311.
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017, 7, 39843.
- Mathieu, O.; Reinders, J.; Caikovski, M.; Smathajitt, C.; Paszkowski, J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 2007, 130, 851–862.
- Zhang, Y.Y.; Fischer, M.; Colot, V.; Bossdorf, O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 2013, 197, 314–322.
- Cortijo, S.; Wardenaar, R.; Colomé-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J.M.; Wincker, P.; et al. Mapping the epigenetic basis of complex traits. Science 2014, 343, 1145–1148.
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408.
- Morgado, L.; Preite, V.; Oplaat, C.; Anava, S.; de Carvalho, J.F.; Rechavi, O.; Johannes, F.; Verhoeven, K.J.F. Small RNAs reflect grandparental environments in apomictic dandelion. Mol. Biol. Evol. 2017, 34, 2035–2040.
- Kuhlmann, M.; Finke, A.; Mascher, M.; Mette, M.F. DNA methylation maintenance consolidates RNA-directed DNA methylation and transcriptional gene silencing over generations in Arabidopsis thaliana. Plant J. 2014, 80, 269–281.
- Wibowo, A.; Becker, C.; Marconi, G.; Durr, J.; Price, J.; Hagmann, J.; Papareddy, R.; Putra, H.; Kageyama, J.; Becker, J.; et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 2016, 5, e13546.
- Ganguly, D.R.; Crisp, P.A.; Eichten, S.R.; Pogson, B.J. The Arabidopsis DNA methylome is stable under transgenerational drought stress. Plant Physiol. 2017, 175, 1893–1912.
- Van Dooren, T.J.M.; Silveira, A.B.; Gilbault, E.; Jiménez-Gómez, J.M.; Martin, A.; Bach, L.; Tisné, S.; Quadrana, L.; Loudet, O.; Colot, V. Mild drought in the vegetative stage induces phenotypic, gene expression and DNA methylation plasticity in Arabidopsis but no transgenerational effects. J. Exp. Bot. 2020, 71, 3588–3602.
- Braun, R.E. Packaging paternal chromosomes with protamine. Nat. Genet. 2001, 28, 10–12.
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093.
- Feng, X.J.; Li, J.R.; Qi, S.L.; Lin, Q.F.; Jin, J.B.; Hua, X.J. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E8335–E8343.
- Calarco, J.P.; Borges, F.; Donoghue, M.T.; Van Ex, F.; Jullien, P.E.; Lopes, T.; Gardner, R.; Berger, F.; Feijó, J.A.; Becker, J.D.; et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 2012, 151, 194–205.
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109.
- Borg, M.; Berger, F. Chromatin remodelling during male gametophyte development. Plant J. 2015, 83, 177–188.
- Borg, M.; Jacob, Y.; Susaki, D.; LeBlanc, C.; Buendía, D.; Axelsson, E.; Kawashima, T.; Voigt, P.; Boavida, L.; Becker, J.; et al. Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 2020, 22, 621–629.
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 2017, 547, 419–424.
- Grossniklaus, U.; Paro, R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331.




