Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Vassilis Souliotis | -- | 3100 | 2022-10-27 11:31:42 | | | |
| 2 | Camila Xu | Meta information modification | 3100 | 2022-10-31 09:41:37 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Papanikolaou, C.; Rapti, V.; Stellas, D.; Stefanou, D.T.; Syrigos, K.; Pavlakis, G.N.; Souliotis, V.L. DNA Damage Response and COVID-19. Encyclopedia. Available online: https://encyclopedia.pub/entry/31860 (accessed on 16 January 2026).
Papanikolaou C, Rapti V, Stellas D, Stefanou DT, Syrigos K, Pavlakis GN, et al. DNA Damage Response and COVID-19. Encyclopedia. Available at: https://encyclopedia.pub/entry/31860. Accessed January 16, 2026.
Papanikolaou, Christina, Vasiliki Rapti, Dimitris Stellas, Dimitra T. Stefanou, Konstantinos Syrigos, George N. Pavlakis, Vassilis L. Souliotis. "DNA Damage Response and COVID-19" Encyclopedia, https://encyclopedia.pub/entry/31860 (accessed January 16, 2026).
Papanikolaou, C., Rapti, V., Stellas, D., Stefanou, D.T., Syrigos, K., Pavlakis, G.N., & Souliotis, V.L. (2022, October 29). DNA Damage Response and COVID-19. In Encyclopedia. https://encyclopedia.pub/entry/31860
Papanikolaou, Christina, et al. "DNA Damage Response and COVID-19." Encyclopedia. Web. 29 October, 2022.
Copy Citation
COVID-19 is an infectious disease caused by the SARS-CoV-2 coronavirus and characterized by an extremely variable disease course, ranging from asymptomatic cases to severe illness. Our cells develop DNA lesions on a daily basis. These lesions can inhibit basic cellular processes, such as genome replication and transcription, and if they are not repaired properly, they could result in mutations or genome aberrations, thereby posing a threat to the cell or even to the viability of a particular organism.
COVID-19 pandemic
SARS-CoV-2 coronavirus
aberrant immune response
DNA damage response
1. Introduction
Coronavirus Disease 2019 (COVID-19) is an infectious disease caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), a novel coronavirus that emerged in the city of Wuhan, China at the end of 2019. Being highly transmissible, the disease was rapidly spread worldwide and a few months later, in March 2020, the World Health Organization (WHO) declared the COVID-19 outbreak a global pandemic [1]. Unlike other coronaviruses that led to large-scale outbreaks (e.g., the SARS-CoV epidemic occurred in 2002, later eradicated; the MERS epidemic: firstly, reported in 2012, still ongoing), COVID-19 has shaped the human history of the 21st century and continues to pose unprecedented challenges for healthcare systems and socioeconomic structures globally [2]. COVID-19 is characterized by an unpredictable and extremely variable disease course ranging from asymptomatic cases to severe illness that can lead even to death. While upper respiratory symptoms are the most common acute manifestations encountered in the majority of patients, many of them develop interstitial pneumonia that may progress to respiratory failure and acute respiratory distress syndrome (ARDS) requiring mechanical ventilation and admission to the intensive care unit (ICU) [3]. Although SARS-CoV-2 predominantly causes pulmonary disease, a wide spectrum of extra-pulmonary clinical manifestations has been also observed. Literature suggests that any system (hematologic, cardiovascular, renal, gastrointestinal and hepatobiliary, endocrinologic, neurologic, ophthalmologic, dermatologic system) can be affected and the main components of SARS-CoV-2 ability to provoke multiple organ injury are (i) direct virus-induced cytotoxicity in angiotensin-converting enzyme 2 (ACE2) expressing cells, (ii) dysregulation of the renin-angiotensin-aldosterone system resulting from virus-mediated ACE2 downregulation related to virus entry, (iii) immune dysregulation, (iv) endothelial cell injury and thrombo-inflammation, and (v) tissue fibrosis [4].
2. DNA Damage Response and COVID-19
2.1. DNA Repair Mechanisms
Our cells develop DNA lesions on a daily basis. These lesions can inhibit basic cellular processes, such as genome replication and transcription, and if they are not repaired properly, they could result in mutations or genome aberrations, thereby posing a threat to the cell or even to the viability of a particular organism [5]. Several endogenous insults are responsible for forming these DNA lesions, including DNA base mismatch, oxidation, hydrolysis, and alkylation of DNA, as well as exogenous factors, such as ultraviolet and ionizing radiation and several chemical agents [6].
To protect against the genotoxic effects, cells have evolved several genome-protection pathways, collectively termed the DNA damage response network (Figure 1) [7]. DDR is an organized system that includes sensors, mediators, transducers, and effectors that activate various pathways, including DNA repair and cell cycle control. If the unrepaired DNA lesions are above a certain level, apoptosis or mutagenesis are triggered [8].
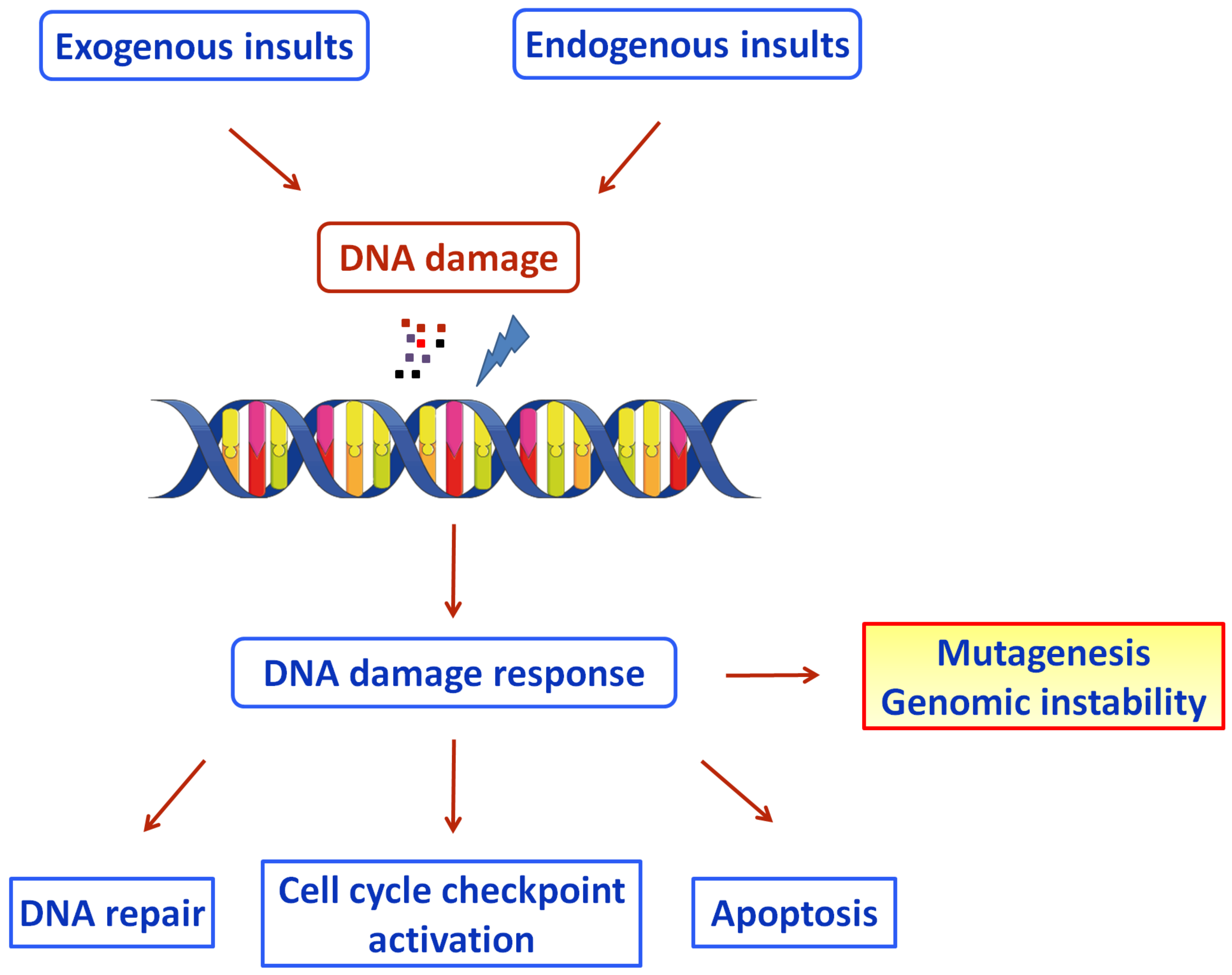
Figure 1. Schematic diagram of the DDR pathways activated by exogenous and endogenous insults (Figure was generated using images assembled from Servier Medical Art, https://smart.servier.com, accessed on 11 August 2022).
At least seven major DNA repair pathways are active throughout the cell cycle, with each one repairing different types of lesions.
- (a)
-
Nucleotide excision repair (NER). This mechanism repairs lesions that disrupt the DNA double-helix, such as bulky base adducts [9]. NER detects helix-distorting base lesions via two sub-pathways with different lesion detection mechanisms: transcription-coupled repair (TCR), which identifies lesions that inhibit transcription, and global-genome repair (GGR), which removes lesions throughout the genome.
- (b)
-
Base excision repair (BER). This is a commonly used DNA repair process that identifies and repairs damaged DNA bases that do not alter the structure of the DNA helix. The cell uses BER to repair abnormal DNA bases, simple base-adducts, oxidative DNA damage and single-strand breaks (SSBs) [10]. There are two BER sub-pathways: the short-patch and the long-patch pathway. The activation of one or both of these two BER sub-pathways is determined by the origin of the damage and the cell cycle phase in which the damage occurs.
- (c)
-
Mismatch repair (MMR). This pathway eliminates base substitution and insertion/deletion mismatches that occur when replication errors escape DNA polymerases’ proofreading function [11].
- (d)
-
Homologous recombination repair (HRR). This is an error-free DSB repair mechanism that works throughout the S and G2 phases of the cell cycle to find a sister chromatid, which serves as a template to direct the repair of the damaged sequence [12].
- (e)
-
Non-homologous end-joining (NHEJ). This mechanism repairs radiation- or chemically-induced double-strand breaks (DSBs), as well as intermediates of the V(D)J recombination and class-switch recombination (CSR) processes [13][14][15]. It is prone to errors and can function at any stage of the cell cycle. There are two subtypes of NHEJ: the canonical (c-NHEJ) and the alternative non-homologous end-joining (alt-NHEJ).
- (f)
-
Interstrand cross-link (ICL) repair. This pathway repairs cross-links between the two strands of DNA, a critical event that usually results in cell cycle and replication arrest and eventually cell death [16]. In non-replicating cells, ICL repair is mediated by the NER mechanism, while in the S phase it is coupled to DNA replication and depends on the homologous recombination machinery [17].
- (g)
-
Direct repair pathway. The only protein that is implicated in this mechanism is the O6-methylguanine-DNA methyltransferase (MGMT), which removes alkyl groups from the O6 position of guanine to a cysteine residue on itself and undergoes the degradation process [18].
2.2. COVID-19 and DDR
During the past few years, a wealth of information has been accumulated, shedding light on interactions between viral infections and the activation of DDR-related pathways [19]. For example, Chambers and colleagues [20] reported that the MMR pathway, a critical component of the DDR network, is required for the cellular anti-influenza A virus (IAV) response and controls the cellular fate following viral infection. Previous studies have shown that IAV infection leads to the death of infected cells through various cell death pathways, such as necrosis, necroptosis and pyroptosis, thus promoting effective virus clearance [21][22]. In addition to the IAV-induced death of infected cells, immune cells can effectively recognize and clear infected cells from the host, thus resulting in viral clearance from the host [23]. Interestingly, although IAV infection typically decreases cells’ MMR capacity, a subset of respiratory epithelial cells, named club cells, are remarkably capable of maintaining high levels of MMR activity [20]. Club cells’ increased MMR capacity efficiently removes the virus-induced oxidative DNA damage, thus allowing the transcriptional activation of antiviral genes, which probably aids in viral eradication and cell survival. In vivo, this has significant clinical implications because the loss of MMR activity reduced cell survival and exacerbated viral illness. In fact, Haque and colleagues [24] reported that a cancer patient with hereditary nonpolyposis colorectal cancer, a syndrome characterized by defective MMR, tested positive for SARS-CoV-2 for at least 54 days after the diagnosis of COVID-19. The authors proposed a connection between a deficient MMR mechanism and protracted viral shedding after SARS-CoV-2 infection, where the host repair system is harmed as a result of the virus-induced oxidative DNA damage and the impaired MMR.
Poly (ADP)-ribose polymerase (PARP) enzymes are a family of proteins that have been extensively investigated in many human diseases, including cancer, disorders of the central nervous system and RNA viral pathology [25]. Although the poly (ADP-ribosylating) (PARylating) PARPs catalyze the formation of branched or linear chains of ADP-ribose moieties and mainly function in the cellular response to DNA damage, several noncanonical mono(ADP-ribosylating) (MARylating) PARPs that modify their target proteins by the addition of a single ADP-ribose moiety, are implicated in cellular antiviral responses [26]. Interestingly, Heer and colleagues [27] have shown that SARS-CoV-2 infection induces MARylating PARPs, such as PARP7, PARP10, PARP12, and PARP14, and up-regulates the expression of genes that are encoding enzymes for salvage nicotinamide adenine dinucleotide (NAD) synthesis from nicotinamide and nicotinamide riboside, while down-regulating other NAD biosynthetic pathways. Importantly, PARP inhibitors had advantageous effects on SARS-CoV-2 infection by blocking the overactivation of macrophages and the cytokine storm that follows [28][29], as well as by preventing cell death [30]. Other studies have also shown that PARP inhibitors showed a protective role against the risk of COVID-19 progression in patients with cardiovascular, central nervous system, and metabolic diseases [31][32].
Furthermore, SARS-CoV-2 infection activated the DDR network in Vero E6, an African green monkey kidney cell line [33]. In that study, virus-infected Vero E6 cells exhibited (a) transcriptional upregulation of the Ataxia telangiectasia and Rad3-related (ATR) protein, (b) increased phosphorylation of Chk1 at serines 317 (S317) and 345 (S345), (c) increased phosphorylation of histone H2AX at serine 139 (S139; γH2AX), (d) decreased expression of the telomeric repeat-binding factor 2 (TRF2) subunit of the Shelterin system, a protein complex that plays a crucial role in telomere protection, while its absence results in DDR activation and the processing of chromosome ends by the DNA repair pathways [34], and (e) decreased telomere lengths. These findings suggest that SARS-CoV-2 affects telomere length, through the decreased expression of the TRF2 protein, thus triggering the DDR network mediated by the ATR signaling pathway. In addition, Sepe and colleagues [35] have shown that during aging the expression of the virus’ cell receptor ACE2 increased in mice and human lungs. They also reported that this increase was dependent on the DDR network, since both (a) the inhibition of the ATM kinase activity, resulting in the global DDR inhibition, and (b) the telomeric DDR inhibition through specific antisense oligonucleotides, prevented the upregulation of ACE2 following telomere shortening or the induction of DNA damage. Together, these results suggest that during aging, telomeric shortening or DNA damage activates the DDR network resulting in the upregulation of ACE2 and making older people more susceptible to SARS-CoV-2 infection.
In another study, the authors claim that SARS-CoV-2 infection of cells expressing high levels of ACE2, the SARS-CoV-2 spike protein induced the formation of syncytia and the generation of micronuclei due to DNA damage [36]. Interestingly, the authors reported that the formation of DNA damage within these syncytial micronuclei triggers the DDR network and the cGAS-STING-IFN signaling, through the recruitment of the γH2Ax and the cGAS proteins, thus resulting in cellular catastrophe and aberrant immune activation.
A recent study shows that exposure of Poecilia reticulata (a widely distributed tropical fish) adult fish to fragments of the SARS-CoV-2 spike protein induced genomic instability, DNA damage in circulating erythrocytes and induction of oxidative stress marked by increased levels of the antioxidant enzymes superoxide dismutase (SOD) and catalase (CAT) and accompanied by high levels of malondialdehyde (MDA), reactive oxygen species (ROS), and hydrogen peroxide (H2O2) [37].
Notably, previous studies have shown that NOX4-derived oxidative stress plays a crucial role in influenza virus proliferation [38]. Indeed, it has been demonstrated that lung epithelial cells infected with the influenza A virus experience a brief rise in intracellular ROS as a necessary phase in the development of the virus life cycle. Through this process, the p38 and ERK1-2 MAPK pathways are activated, which in turn promotes the nuclear export of viral ribonucleoprotein (vRNP), a crucial step in viral assembly and release. Remarkably, the NOX4 oxidase was the major contributor to the virus-induced oxidative stress and the enhancement of viral replication in murine primary airway epithelial cells and human lung cancer cell lines. Indeed, it was found that the expression of NOX4 was increased during cell infection, while inhibition of NOX4 activity blocked ROS increase, the phosphorylation of MAPK, the nuclear export of the vRNP and the viral release [38].
In addition, Garcia and colleagues [39] shed light on the interactions between COVID-19 and the DDR pathway and suggested DDR-associated kinase inhibitors as potent blockers of SARS-CoV-2 replication. Indeed, the authors screened a library of pharmacological compounds to find antiviral medicines that are specific to SARS-CoV-2 and found that virus cytopathic impact in human epithelial cells was inhibited by 34 of 430 protein kinase inhibitors that are in different phases of clinical trials. For example, berzosertib, a selective inhibitor of serine/threonine-protein kinase ATR, that is already in phase 2 clinical trials for solid tumors [40], prevented SARS-CoV-2 replication at the post-entry stage and had significant antiviral action in various cell types.
Importantly, a recent study has shown how COVID-19 damages the heart, opening the opportunity for new COVID-19 treatments [41]. In that study, the authors investigated the transcriptome landscape of cardiac tissues collected from SARS-CoV-2 infected patients and controls. Transcriptomics analysis showed upregulation of DNA damage and repair-related genes in the cardiac tissues of COVID-19 patients. In addition, the presence of DNA damage in the same tissues of SARS-CoV-2 patients was further confirmed using γH2AX immunostaining, an established methodology for detecting DNA damage.
2.3. COVID-19 and Oxidative Stress
Oxidative stress is defined as a dangerous state caused by the imbalance between the production and the accumulation of ROS [42]. ROS are highly reactive molecules that trigger rapid chain reactions and cause oxidative damage to macromolecules, such as lipids, proteins, carbohydrates, and nucleic acids, thus affecting various cellular functions. Previous data have shown that increased levels of oxidative stress participate in the onset and progression of many diseases, such as cancer and autoimmunity [43]. On the other hand, very low levels of oxidative stress result in the induction of reductive stress and the occurrence of pathologies ranging from cancer to cardiomyopathy [44].
A growing number of studies have shown that oxidative stress plays a key role in viral infections, such as SARS-CoV-2 [45][46][47]. Indeed, a primary characteristic of viral infection is an imbalance of redox equilibrium in the body [48]. By creating an excess of ROS and a shortage of reduced glutathione (GSH), the virus manipulates the host cell machinery to put the cell into an oxidative stress state, which creates favorable conditions for viral reproduction. Interestingly, the excess reactive oxygen/nitrogen species (RONS) synthesis and the abnormal cellular antioxidant-oxidant balance have been implicated in the pathogenesis of respiratory viral infections, such as SARS-CoV-2 [45].
Other studies suggested that oxidation of thiols to disulfides, because of oxidative stress, might boost SARS-CoV-2 spike protein affinity for the ACE2 receptor that is responsible for the degradation of the vasoconstrictor Angiotensin II (Ang II) to the vasodilator Angiotensin 1–7 (Ang 1–7) and so increase the severity of SARS-CoV-2 infection [49][50]. Because SARS-CoV-2 binding to the ACE2 receptor reduces the enzyme’s catalytic activity, i.e., the conversion of Ang II to Ang 1–7, the nicotinamide dinucleotide phosphate (reduced form, NADPH) oxidase activity may also rise in SARS-CoV-2 infected patients, resulting in an increase of oxidative stress [51]. Moreover, lower levels of the antioxidant glutathione enhance cellular oxidative stress, which is linked to a variety of diseases and immunological dysfunctions that increase viral infection susceptibility, such as uncontrolled SARS-CoV-2 infection [52]. Researchers also have to mention that uncontrolled replication causes oxidative damage in the lungs, increasing viral load and consequently the severity of the virus infection [53]. Moreover, since the membrane antioxidant vitamin D has been reported to improve immunity and protect against respiratory illness, a recent study proposed that there is a connection between vitamin D levels and COVID-19 susceptibility [54][55].
Since oxidative stress might directly or indirectly affect the progression and outcome of SARS-CoV-2 infection, it is an emerging target in the battle against viral infection. Indeed, many studies have reported the use of antioxidants, such as N-acetylcysteine (NAC), GSH, polyphenols, and selenium, in the treatment of viral infections [56][57]. For example, a recent study has shown that NAC, due to its participation in the synthesis of glutathione, by boosting T cell response, preventing the depletion of the T cells and reducing inflammation, could be a promising medication to treat COVID-19 infection [58][59]. Other studies demonstrated that the in vitro or in vivo administration of GSH derivatives inhibited Sendai virus and Herpes Simplex Virus 1 (HSV-1) replication, without inducing toxic effects [60][61]. Moreover, previous data have shown that polyphenol components derived from pistachios kernels (the raw kernels of the pistachio nut) exhibited antiviral effects, with resveratrol, a stilbene derived from a variety of plants, being the best anti-HSV nutraceutical agent [62][63]. In addition, selenium-based nanoparticles have emerged as a promising approach in the treatment of influenza [64].
Importantly, in a recent study, the authors measured redox biomarkers and DNA damage levels for 14 days in hospitalized COVID-19 patients. Maximal levels of malondialdehyde, a biomarker of lipid peroxidation and oxidative stress, were observed at the time of hospitalization, rapidly dropping during the time-course analyzed, while 8-hydroxy-2′-deoxyguanosine (8-OHdG) levels, an important byproduct of oxidative DNA damage, peaked at 7 days after hospitalization [65]. Another study evaluated the presence of guanine oxidized species in COVID-19 hospitalized patients [66]. The authors reported that the levels of the DNA and RNA guanine oxidized species were higher in the serum of non-surviving COVID-19 patients than in those that survived, suggesting that oxidative DNA damage could be a predicting factor of death by COVID-19.
2.4. SARS-CoV-2 Vaccination, the Immune System and the DDR Network
The DDR network and the immune system are the main mechanisms that act together favoring the proper function of various organisms [8]. Indeed, several studies have shown that activated DDR induces immune responses, usually via a cGAS/STING-mediated pathway [67][68][69], while the activated immune system induces the DDR network through the generation of oxidative stress and the resulting DNA damage [70][71]. To further explore the interplay between these two systems, Ntouros and colleagues [72] investigated the effect of an acute immune challenge on the DDR system, using SARS-CoV-2 vaccination as an in vivo model of acute inflammation. They found that 24 h after SARS-CoV-2 vaccination (Comirnaty, Pfizer-BioNTech), peripheral blood mononuclear cells (PBMCs) of healthy individuals showed a transient increase of type I IFN, combined with elevated oxidative stress and accumulation of DNA damage; vaccination did not influence the DNA repair capacity of PBMCs. All these parameters resumed regular levels a few days later. Collectively, these data show that SARS-CoV-2 vaccination, as an acute immune stimulant, successfully triggers the DDR network. Moreover, the cytokine profile of the vaccinated individuals reveals a distinct interleukin 15, interferon gamma and IP10/CXCL10 signature, which correlates with effective immune activation [73].
A growing number of studies have shown that older people are characterized by decreased antibody response to SARS-CoV-2 vaccination [74]. To elucidate the link between aging and the response to SARS-CoV-2 vaccination, a recent report analyzed oxidative stress and accumulation of DNA damage in aged individuals before and after vaccination [75]. They found that after SARS-CoV-2 vaccination (Comirnaty, Pfizer-BioNTech, New York, NY 10017, USA), individual titers of anti-Spike-Receptor Binding Domain (S-RBD)-IgG antibodies and the neutralizing capacity of circulating anti-SARS-CoV-2 antibodies inversely correlated with the corresponding pre-vaccination oxidative stress status and the DNA damage levels observed in PBMCs. Together, these results suggest that humoral immune responses to SARS-CoV-2 vaccination may be weaker when immune cells are under oxidative and/or genotoxic stress, conditions that are common in the elderly.
References
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160.
- Hu, T.; Liu, Y.; Zhao, M.; Zhuang, Q.; Xu, L.; He, Q. A comparison of COVID-19, SARS and MERS. PeerJ 2020, 8, e9725.
- Boechat, J.L.; Chora, I.; Morais, A.; Delgado, L. The immune response to SARS-CoV-2 and COVID-19 immunopathology—Current perspectives. Pulmonology 2021, 27, 423–437.
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032.
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078.
- Hakem, R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008, 27, 589–605.
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 14, 739–745.
- Pateras, I.S.; Havaki, S.; Nikitopoulou, X.; Vougas, K.; Townsend, P.A.; Panayiotidis, M.I.; Georgakilas, A.G.; Gorgoulis, V.G. The DNA damage response and immune signaling alliance: Is it good or bad? Nature decides when and where. Pharmacol. Ther. 2015, 154, 36–56.
- Shuck, S.C.; Short, E.A.; Turchi, J.J. Eukaryotic nucleotide excision repair: From understanding mechanisms to influencing biology. Cell Res. 2008, 18, 64–72.
- Krokan, H.E.; Bjørås, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, 012583.
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346.
- Helleday, T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 2010, 6, 955–960.
- Chakraborty, A.; Tapryal, N.; Venkova, T.; Horikoshi, N.; Pandita, R.K.; Sarker, A.H.; Sarkar, P.S.; Pandita, T.K.; Hazra, T.K. Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 2016, 7, 13049.
- Iliakis, G.; Murmann, T.; Soni, A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 793, 166–175.
- Lieber, M.R. The mechanism of human nonhomologous DNA end-joining. J. Biol. Chem. 2008, 283, 1–5.
- Hashimoto, S.; Anai, H.; Hanada, K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016, 38, 9.
- Wang, Y.; Leung, J.W.; Jiang, Y.; Lowery, M.G.; Do, H.; Vasquez, K.M.; Chen, J.; Wang, W.; Li, L. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol. Cell 2013, 49, 997–1009.
- Hiddinga, B.I.; Pauwels, P.; Janssens, A.; van Meerbeeck, J.P. O6-Methylguanine-DNA methyltransferase (MGMT): A drugable target in lung cancer? Lung Cancer 2017, 107, 91–99.
- Ryan, E.L.; Hollingworth, R.; Grand, R.J. Activation of the DNA damage response by RNA viruses. Biomolecules 2016, 6, 2.
- Chambers, B.S.; Heaton, B.E.; Rausch, K.; Dumm, R.E.; Hamilton, J.R.; Cherry, S.; Heaton, N.S. DNA mismatch repair is required for the host innate response and controls cellular fate after influenza virus infection. Nat. Microbiol. 2019, 4, 1964–1977.
- Shubina, M.; Tummers, B.; Boyd, D.F.; Zhang, T.; Yin, C.; Gautam, A.; Guo, X.J.; Rodriguez, D.A.; Kaiser, W.J.; Vogel, P.; et al. Necroptosis restricts influenza A virus as a stand-alone cell death mechanism. J. Exp. Med. 2020, 217, e20191259.
- Downey, J.; Pernet, E.; Coulombe, F.; Divangahi, M. Dissecting host cell death programs in the pathogenesis of influenza. Microbes Infect. 2018, 20, 560–569.
- Van de Sandt, C.E.; Kreijtz, J.H.; Rimmelzwaan, G.F. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 2012, 4, 1438–1476.
- Haque, F.; Lillie, P.; Haque, F.; Maraveyas, A. Deficient DNA mismatch repair and persistence of SARS-CoV-2 RNA shedding: A case report of hereditary nonpolyposis colorectal cancer with COVID-19 infection. BMC Infect. Dis. 2021, 21, 854.
- Dash, S.; Dash, C.; Pandhare, J. Therapeutic Significance of microRNA-Mediated Regulation of PARP-1 in SARS-CoV-2 Infection. Non-Coding RNA 2021, 7, 60.
- Challa, S.; Stokes, M.S.; Kraus, W.L. MARTs and MARylation in the Cytosol: Biological Functions, Mechanisms of Action, and Therapeutic Potential. Cells 2021, 10, 313.
- Heer, C.D.; Sanderson, D.J.; Voth, L.S.; Alhammad, Y.M.O.; Schmidt, M.S.; Trammell, S.A.J.; Perlman, S.; Cohen, M.S.; Fehr, A.R.; Brenner, C. Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity. J. Biol. Chem. 2020, 295, 17986–17996.
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034.
- Shi, Y.; Wang, Y.; Shao, C.; Huang, J.; Gan, J.; Huang, X.; Bucci, E.; Piacentini, M.; Ippolito, G.; Melino, G. COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 2020, 27, 1451–1454.
- Fehr, A.R.; Singh, S.A.; Kerr, C.M.; Mukai, S.; Higashi, H.; Aikawa, M. The impact of PARPs and ADP-ribosylation on inflammation and host–pathogen interactions. Genes Dev. 2020, 34, 341–359.
- Bai, P. Biology of poly (ADP-ribose) polymerases: The factotums of cell maintenance. Mol. Cell 2015, 58, 947–958.
- Fatokun, A.A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016.
- Victor, J.; Deutsch, J.; Whitaker, A.; Lamkin, E.N.; March, A.; Zhou, P.; Botten, J.W.; Chatterjee, N. SARS-CoV-2 triggers DNA damage response in Vero E6 cells. Biochem. Biophys. Res. Commun. 2021, 579, 141–145.
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110.
- Sepe, S.; Rossiello, F.; Cancila, V.; Iannelli, F.; Matti, V.; Cicio, G.; Cabrini, M.; Marinelli, E.; Alabi, B.R.; di Lillo, A.; et al. DNA damage response at telomeres boosts the transcription of SARS-CoV-2 receptor ACE2 during aging. EMBO Rep. 2022, 23, 53658.
- Ren, H.; Ma, C.; Peng, H.; Zhang, B.; Zhou, L.; Su, Y.; Gao, X.; Huang, H. Micronucleus production, activation of DNA damage response and cGAS-STING signaling in syncytia induced by SARS-CoV-2 infection. Biol. Direct. 2021, 16, 20.
- Gonçalves, S.O.; Luz, T.M.D.; Silva, A.M.; de Souza, S.S.; Montalvão, M.F.; Guimarães, A.T.B.; Ahmed, M.A.I.; Araújo, A.P.D.C.; Karthi, S.; Malafaia, G. Can spike fragments of SARS-CoV-2 induce genomic instability and DNA damage in the guppy, Poecilia reticulate? An unexpected effect of the COVID-19 pandemic. Sci. Total Environ. 2022, 825, 153988.
- Amatore, D.; Sgarbanti, R.; Aquilano, K.; Baldelli, S.; Limongi, D.; Civitelli, L.; Nencioni, L.; Garaci, E.; Ciriolo, M.R.; Palamara, A.T. Influenza virus replication in lung epithelial cells depends on redox-sensitive pathways activated by NOX4-derived ROS. Cell Microbiol. 2015, 17, 131–145.
- Garcia, G., Jr.; Sharma, A.; Ramaiah, A.; Sen, C.; Purkayastha, A.; Kohn, D.B.; Parcells, M.S.; Beck, S.; Kim, H.; Bakowski, M.A.; et al. Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication. Cell Rep. 2021, 35, 108940.
- Konstantinopoulos, P.A.; Cheng, S.C.; Wahner Hendrickson, A.E.; Penson, R.T.; Schumer, S.T.; Doyle, L.A.; Lee, E.K.; Kohn, E.C.; Duska, L.R.; Crispens, M.A.; et al. Berzosertib plus gemcitabine versus gemcitabine alone in platinum-resistant high-grade serous ovarian cancer: A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020, 21, 957–968.
- Kulasinghe, A.; Liu, N.; Tan, C.W.; Monkman, J.; Sinclair, J.E.; Bhuva, D.D.; Godbolt, D.; Pan, L.; Nam, A.; Sadeghirad, H.; et al. Transcriptomic profiling of cardiac tissues from SARS-CoV-2 patients identifies DNA damage. Immunology 2022, in press.
- Souliotis, V.L.; Vlachogiannis, N.I.; Pappa, M.; Argyriou, A.; Ntouros, P.A.; Sfikakis, P.P. DNA Damage Response and Oxidative Stress in Systemic Autoimmunity. Int. J. Mol. Sci. 2019, 21, 55.
- Zuo, L.; Prather, E.R.; Stetskiv, M.; Garrison, D.E.; Meade, J.R.; Peace, T.I.; Zhou, T. Inflammaging and Oxidative Stress in Human Diseases: From Molecular Mechanisms to Novel Treatments. Int. J. Mol. Sci. 2019, 20, 4472.
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496.
- Ntyonga-Pono, M.P. COVID-19 infection and oxidative stress: An under-explored approach for prevention and treatment? Pan. Afr. Med. J. 2020, 35, 12.
- Schönrich, G.; Raftery, M.J.; Samstag, Y. Devilishly radical NETwork in COVID-19: Oxidative stress, neutrophil extracellular traps (NETs), and T cell suppression. Adv. Biol. Regul. 2020, 77, 100741.
- Laforge, M.; Elbim, C.; Frère, C.; Hémadi, M.; Massaad, C.; Nuss, P.; Benoliel, J.J.; Becker, C. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 2020, 20, 515–516.
- Checconi, P.; De Angelis, M.; Marcocci, M.E.; Fraternale, A.; Magnani, M.; Palamara, A.T.; Nencioni, L. Redox-Modulating Agents in the Treatment of Viral Infections. Int. J. Mol. Sci. 2020, 21, 4084.
- Hati, S.; Bhattacharyya, S. Impact of Thiol-Disulfide Balance on the Binding of COVID-19 Spike Protein with Angiotensin-Converting Enzyme 2 Receptor. ACS Omega 2020, 5, 16292–16298.
- Busse, L.W.; Chow, J.H.; McCurdy, M.T.; Khanna, A.K. COVID-19 and the RAAS-a potential role for angiotensin II? Crit. Care 2020, 24, 136.
- Suhail, S.; Zajac, J.; Fossum, C.; Lowater, H.; McCracken, C.; Severson, N.; Laatsch, B.; Narkiewicz-Jodko, A.; Johnson, B.; Liebau, J.; et al. Role of Oxidative Stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) Infection: A Review. Protein J. 2020, 39, 644–656.
- Fraternale, A.; Paoletti, M.F.; Casabianca, A.; Oiry, J.; Clayette, P.; Vogel, J.U.; Cinatl, J.J.; Palamara, A.T.; Sgarbanti, R.; Garaci, E.; et al. Antiviral and immunomodulatory properties of new pro-glutathione (GSH) molecules. Curr. Med. Chem. 2006, 13, 1749–1755.
- Polonikov, A. Endogenous Deficiency of Glutathione as the Most Likely Cause of Serious Manifestations and Death in COVID-19 Patients. ACS Infect. Dis. 2020, 6, 1558–1562.
- Kalvandi, F.; Azarbayjani, M.A.; Azizbeigi, R.; Azizbeigi, K. Elastic resistance training is more effective than vitamin D3 supplementation in reducing oxidative stress and strengthen antioxidant enzymes in healthy men. Eur. J. Clin. Nutr. 2022, 76, 610–615.
- Razdan, K.; Singh, K.; Singh, D. Vitamin D Levels and COVID-19 Susceptibility: Is there any Correlation? Med. Drug Discov. 2020, 7, 100051.
- Saso, L.; Firuzi, O. Pharmacological applications of antioxidants: Lights and shadows. Curr. Drug Targets 2014, 15, 1177–1799.
- Sgarbanti, R.; Amatore, D.; Celestino, I.; Marcocci, M.E.; Fraternale, A.; Ciriolo, M.R.; Magnani, M.; Saladino, R.; Garaci, E.; Palamara, A.T.; et al. Intracellular redox state as target for anti-influenza therapy: Are antioxidants always effective? Curr. Top. Med. Chem. 2014, 14, 2529–2541.
- De Flora, S.; Balansky, R.; La Maestra, S. Rationale for the use of N-acetylcysteine in both prevention and adjuvant therapy of COVID-19. FASEB J. 2020, 34, 13185–13193.
- Poe, F.L.; Corn, J. N-Acetylcysteine: A potential therapeutic agent for SARS-CoV-2. Med. Hypotheses 2020, 143, 109862.
- Palamara, A.T.; Brandi, G.; Rossi, L.; Millo, E.; Benatti, U.; Nencioni, L.; Iuvara, A.; Garaci, E.; Magnani, M. New synthetic glutathione derivatives with increased antiviral activities. Antivir. Chem. Chemother. 2004, 15, 83–91.
- Sgarbanti, R.; Nencioni, L.; Amatore, D.; Coluccio, P.; Fraternale, A.; Sale, P.; Mammola, C.L.; Carpino, G.; Gaudio, E.; Magnani, M.; et al. Redox regulation of the influenza hemagglutinin maturation process: A new cell-mediated strategy for anti-influenza therapy. Antioxid. Redox. Signal. 2011, 15, 593–606.
- Musarra-Pizzo, M.; Pennisi, R.; Ben-Amor, I.; Smeriglio, A.; Mandalari, G.; Sciortino, M.T. In vitro anti-HSV-1 activity of polyphenol-rich extracts and pure polyphenol compounds derived from pistachios kernels (Pistacia vera L.). Plants 2020, 9, 267.
- Annunziata, G.; Maisto, M.; Schisano, C.; Ciampaglia, R.; Narciso, V.; Tenore, G.C.; Novellino, E. Resveratrol as a novel anti-herpes simplex virus nutraceutical agent: An overview. Viruses 2018, 10, 473.
- Li, Y.; Lin, Z.; Guo, M.; Zhao, M.; Xia, Y.; Wang, C.; Xu, T.; Zhu, B. Inhibition of H1N1 influenza virus-induced apoptosis by functionalized selenium nanoparticles with amantadine through ROS-mediated AKT signaling pathways. Int. J. Nanomed. 2018, 13, 2005–2016.
- Kosanovic, T.; Sagic, D.; Djukic, V.; Pljesa-Ercegovac, M.; Savic-Radojevic, A.; Bukumiric, Z.; Lalosevic, M.; Djordjevic, M.; Coric, V.; Simic, T. Time Course of Redox Biomarkers in COVID-19 Pneumonia: Relation with Inflammatory, Multiorgan Impairment Biomarkers and CT Findings. Antioxidants 2021, 10, 1126.
- Lorente, L.; Martín, M.M.; González-Rivero, A.F.; Pérez-Cejas, A.; Cáceres, J.J.; Perez, A.; Ramos-Gómez, L.; Solé-Violán, J.; Marcos, Y.; Ramos, J.A.; et al. DNA and RNA Oxidative Damage and Mortality of Patients with COVID-19. Am. J. Med. Sci. 2021, 361, 585–590.
- Härtlova, A.; Erttmann, S.F.; Raffi, F.A.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kröger, A.; Nilsson, J.A.; et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 2015, 42, 332–343.
- Günther, C.; Kind, B.; Reijns, M.A.; Berndt, N.; Martinez-Bueno, M.; Wolf, C.; Tüngler, V.; Chara, O.; Lee, Y.A.; Hübner, N.; et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Investig. 2015, 125, 413–424.
- Gehrke, N.; Mertens, C.; Zillinger, T.; Wenzel, J.; Bald, T.; Zahn, S.; Tüting, T.; Hartmann, G.; Barchet, W. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 2013, 39, 482–495.
- Jaiswal, M.; LaRussom, N.F.; Burgart, L.J.; Gores, G.J. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 2000, 60, 184–190.
- Meira, L.B.; Bugni, J.M.; Green, S.L.; Lee, C.W.; Pang, B.; Borenshtein, D.; Rickman, B.H.; Rogers, A.B.; Moroski-Erkul, C.A.; McFaline, J.L.; et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Investig. 2008, 118, 2516–2525.
- Ntouros, P.A.; Vlachogiannis, N.I.; Pappa, M.; Nezos, A.; Mavragani, C.P.; Tektonidou, M.G.; Souliotis, V.L.; Sfikakis, P.P. Effective DNA damage response after acute but not chronic immune challenge: SARS-CoV-2 vaccine versus Systemic Lupus Erythematosus. Clin. Immunol. 2021, 229, 108765.
- Bergamaschi, C.; Terpos, E.; Rosati, M.; Angel, M.; Bear, J.; Stellas, D.; Karaliota, S.; Apostolakou, F.; Bagratuni, T.; Patseas, D.; et al. Systemic IL-15, IFN-γ, and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell Rep. 2021, 36, 109504.
- Hyams, C.; Marlow, R.; Maseko, Z.; King, J.; Ward, L.; Fox, K.; Heath, R.; Tuner, A.; Friedrich, Z.; Morrison, L.; et al. Effectiveness of BNT162b2 and ChAdOx1 nCoV-19 COVID-19 vaccination at preventing hospitalisations in people aged at least 80 years: A test-negative, case-control study. Lancet Infect. Dis. 2021, 21, 1539–1548.
- Ntouros, P.A.; Kravvariti, E.; Vlachogiannis, N.I.; Pappa, M.; Trougakos, I.P.; Terpos, E.; Tektonidou, M.G.; Souliotis, V.L.; Sfikakis, P.P. Oxidative stress and endogenous DNA damage in blood mononuclear cells may predict anti-SARS-CoV-2 antibody titers after vaccination in older adults. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166393.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.1K
Entry Collection:
COVID-19
Revisions:
2 times
(View History)
Update Date:
31 Oct 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




