Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Giorgia Condrò | -- | 3216 | 2022-10-26 16:35:28 | | | |
| 2 | Rita Xu | -15 word(s) | 3201 | 2022-10-27 05:04:12 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Condrò, G.; Guerini, M.; Castello, M.; Perugini, P. Atopic Dermatitis (AD). Encyclopedia. Available online: https://encyclopedia.pub/entry/31460 (accessed on 08 February 2026).
Condrò G, Guerini M, Castello M, Perugini P. Atopic Dermatitis (AD). Encyclopedia. Available at: https://encyclopedia.pub/entry/31460. Accessed February 08, 2026.
Condrò, Giorgia, Marta Guerini, Michela Castello, Paola Perugini. "Atopic Dermatitis (AD)" Encyclopedia, https://encyclopedia.pub/entry/31460 (accessed February 08, 2026).
Condrò, G., Guerini, M., Castello, M., & Perugini, P. (2022, October 26). Atopic Dermatitis (AD). In Encyclopedia. https://encyclopedia.pub/entry/31460
Condrò, Giorgia, et al. "Atopic Dermatitis (AD)." Encyclopedia. Web. 26 October, 2022.
Copy Citation
The skin harbors a huge number of different microorganisms such as bacteria, fungi and viruses, and it acts as a protective shield to prevent the invasion of pathogens and to maintain the health of the commensal microbiota. Several studies, in fact, have shown the importance of the skin microbiota for healthy skin. This balance can be altered by intrinsic and extrinsic factors, leading to the development of skin disease, such as acne vulgaris (AV), atopic dermatitis (AD) and rosacea (RS).
skin bioengineering
skin disease
dermatitis
1. Introduction
The skin is an extensive and dynamic system in which different living microorganisms (bacteria, fungi, viruses, and mites) populate not only the surface but also the deeper layers of the epidermis and dermis, as shown in Figure 1. The entirety of the population composes the microbiota [1][2][3]. This term refers to all commensal microorganisms, which are usually harmless [4], present in and on our bodies, such as in the intestine, nose, mucous membranes, scalp and skin.
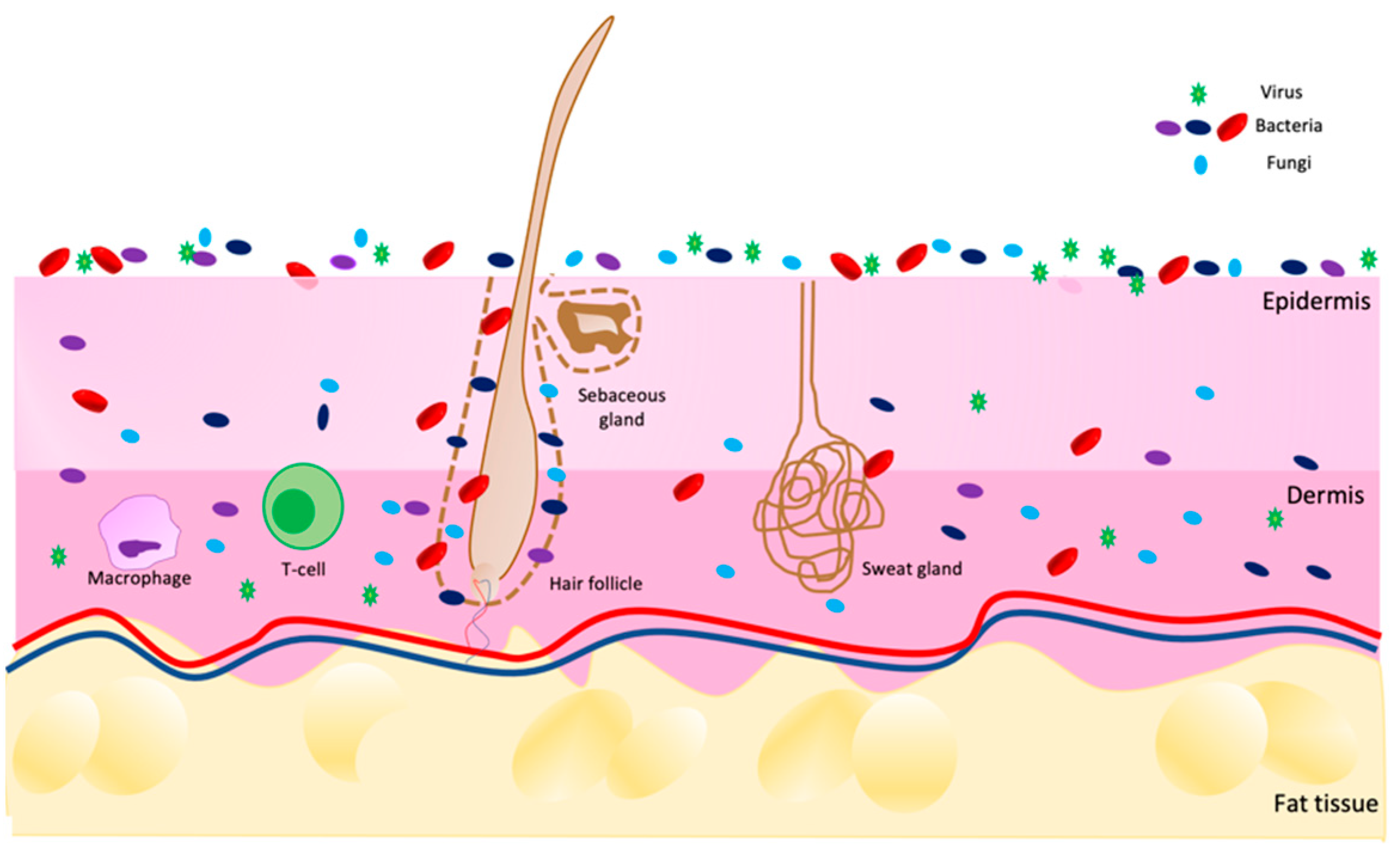
Figure 1. Scheme of skin microbiota distribution on skin in a healthy condition. Image built on Power Point software.
Their function is to defend our body from pathogens and to maintain good health over time [5][6]. To keep a good balance, it is necessary that different microbial communities participate in a symbiotic relationship with the host tissue, conferring it some benefits [7]. In fact, the microbiota includes two types of microorganisms: resident and transient. The first group represents the core of the skin environment, and they help to maintain healthy conditions. In fact, they are neither aggressive nor pathogenic, but they provide some benefits to the host, also preserving the skin barrier. The latter, as the name suggests, do not stabilize permanently, but remain on the skin for only a short period. Under normal conditions, they are not pathogenic.
However, exogenous and endogenous factors can perturb the bacterial flora, leading to changes in the relative abundance of normal commensals, and these could become opportunistic pathogens under the right circumstances [8], contributing to the symptoms [1][2] of diseases such as acne vulgaris (AV), atopic dermatitis (AD) and rosacea (RS) [9][10][11].
The distribution of the skin microbiota is not homogeneous, but it differs according to body areas and to skin characteristics. At the macroscopic level, researchers can subdivide moist, dry, and sebaceous environments. In the sebaceous areas, researchers find lipophilic bacteria such as Cutibacterium, which is able to grow well in anaerobic, lipid-rich regions (for example, the forehead and nose crease). In dry areas, on the other hand, Micrococcus, Streptococcus, and Corynebacterium prevail, such as in the upper buttock area and forearm.
At last, Staphylococcus and Corynebacteria species prefer moist areas, e.g., the armpit, inguinal fold and inner elbow [8][12].
If researchers consider smaller body regions, at the microscopic level (eccrine and apocrine glands, sebaceous glands, and hair follicles), more microbial heterogeneity is present. Cutibacterium species predominate, for example, in sebaceous follicles.
In addition to microbial heterogeneity, bacterial flora also varies from individual to individual by birth type. In the beginning, the fetus in utero is sterile, so the skin microbiota begins to develop at the time of birth. If the childbirth method is cesarean, then the newborn will have a microbial population similar to the mother, and species of Staphylococcus, Corynebacterium, and Cutibacterium will prevail. If, on the other hand, the childbirth is natural, then the baby will have a vaginal microbial set, such as Lactobacillus, Prevotella, and Sneathia [13].
Then, the enrichment of the microbiota continues with the lactation phase, during which the mother’s microorganisms try to reach all areas of the body, including hair follicles and then the scalp, in order to establish a healthy relationship with the skin cells of the host. Adulthood is then the final stage in which everyone will have their own bacterial flora that will not be static, but will change over time. These temporal changes are some other factors affecting interindividual and intraindividual variability. In a study conducted over the course of 4–6 months, it was observed that microbial dynamism was greater in dry environments than in mixed environments.
In addition, other factors that increase the variability of microbial flora are lifestyle, work, race and age. During puberty, young people have greater sebum production, which consequently correlates with a greater presence of lipophilic bacteria such as Cutibacterium.
Another aspect to consider is the environmental impact, including climate, temperature, and UV exposure. Indeed, UVA and UVB radiation are bactericidal [8][12].
Under healthy conditions, the most prevalent bacteria phyla are Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria; in particular, the three most common genera are Corynecateria, Cutibacteria and Staphylococci [13][14][15][16].
Among the commensal bacteria, the most representative are Cutibacterium acnes, Staphylococcus epidermidis and Corynebacterium jeikeium.
Cutibacterium acnes, a Gram-positive anaerobe bacterium, resides in the sebaceous areas of the body, such as the face, neck, hair follicles and sebaceous glands. Other species, such as C. avidum and C. granulosum, are also found, but in smaller quantities. Cutibacterium acnes is considered one of the main commensal bacteria of the skin; in fact, it metabolizes fatty acids with antimicrobial properties and contributes to maintaining the acidic pH of the skin. It also produces bactericides, preventing the growth of yeasts, molds and some Gram-negative pathogens. However, under certain conditions, some subspecies of C. acnes are implicated in acne [15][16].
Staphylococcus epidermidis, which is Gram-positive, belongs to the Firmicutes phylum. It is commonly found on the skin and is considered beneficial: it is able to secrete antimicrobial peptides (AMPs), such as epidermin and epilancin K7, which prevent the colonization of skin pathogens, including Group A Streptococci (S. pyogens) and S. aureus.
Corynebacteria, Gram-positive bacteria (phylum Actinobacteria), colonize moist or sebaceous sites and use lipids or vitamins from sweat to survive.
Members of this family include C. jeikeium; this is a commensal bacterium that, in much the same way as S. epidermidis, produces bacteriocin-like antimicrobial compounds, preventing the colonization of other potentially harmful species. In addition, C. jeikeium also produces superoxide dismutase, an enzyme that protects bacteria from superoxide radicals and simultaneously may prevent oxidative damage at the tissue level [17][18].
The skin microbiota does not include only bacteria but also fungi. Indeed, different studies have suggested that the most present fungi on the skin belong to the Malassezia species, such as M. globosa, M. restricta and M. sympodialis. The distribution of these microbes is body-dependent; for example, M. globosa lives on the back and occiput, whereas M. restricta is instead on the external auditory canal and retroauricular fold. Aspergillus, Rhodotorula, Cryptococcus and Epicoccum are present in other skin areas, such as foot sites [17].
The field of viruses, on the other hand, is not completely known. This may be due to the difficulty of sampling and sequencing them, for example, due to their size [18].
2. Atopic Dermatitis
Atopic dermatitis is a chronic inflammatory skin disease in which the major symptoms are xerosis, dry itchy skin and eczema. In this condition, the skin barrier is compromised, and individuals are more exposed to secondary infections, penetration of allergens, and transepidermal water loss (TEWL) [17][18][19]. Moreover, other characteristics such as inflammation, immune dysregulation and filaggrin mutation can affect AD patients (Figure 2) [19][20][21].
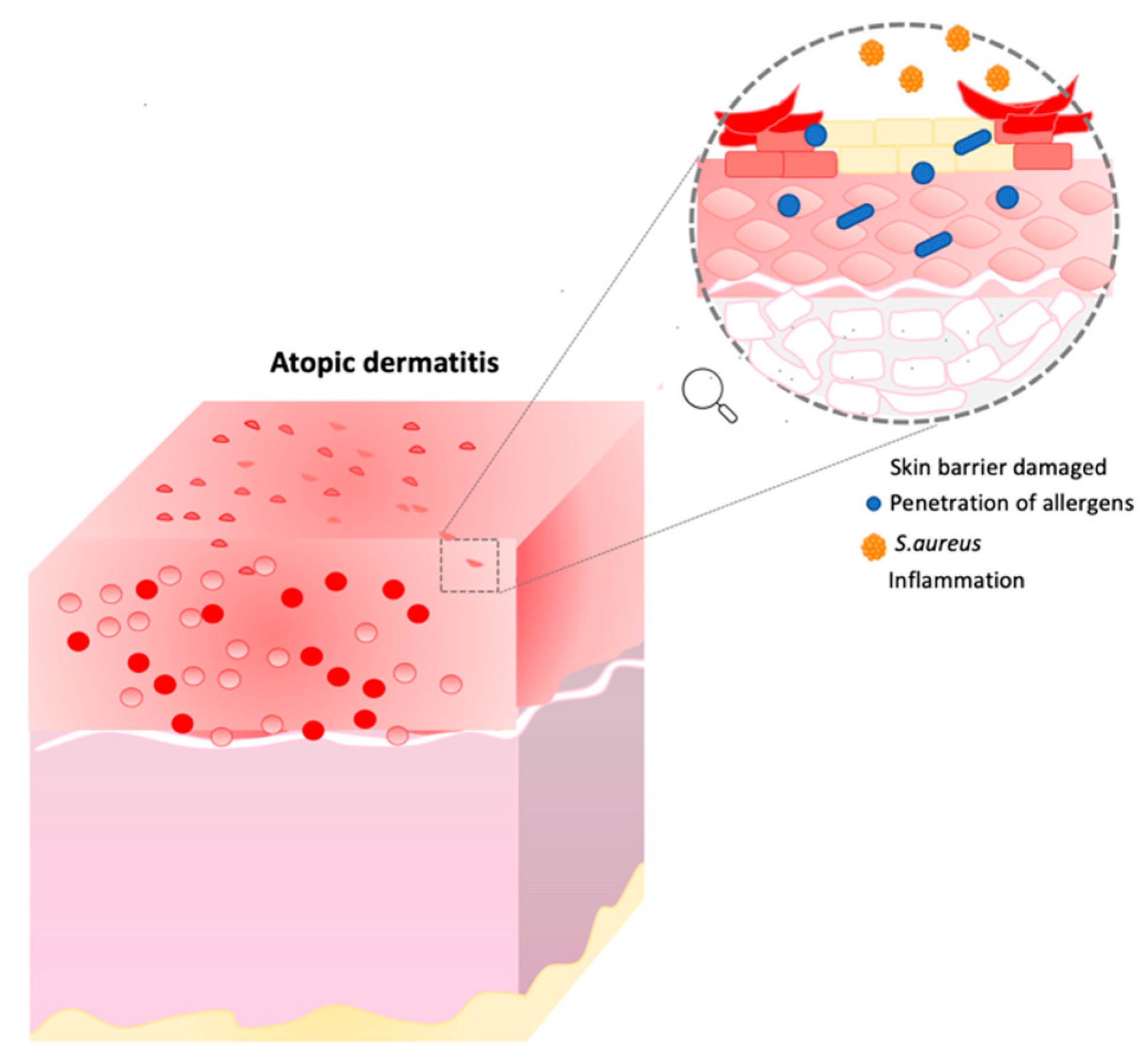
Figure 2. Atopic dermatitis representation. This image describes the consequence of skin barrier damage. Image built on Power Point software.
2.1. Mutation of Filaggrin
Among the causes researchers can find in AD patients, there could be a mutation in the gene (FLG) encoding for the protein filaggrin, localized on the short arm of chromosome 1q21 [22]. Filaggrin is a structural, S100 calcium-binding epidermal stratum corneum (SC) protein. It is involved in the normal SC function in terms of the hydration and maintenance of the skin barrier. It is also responsible for the binding of the keratin filament to produce micro-fibrils. During epidermal differentiation, its insoluble precursor, profilaggrin, is dephosphorylated and becomes more soluble [23]. Then, different proteases cleave the molecule into monomers, forming the final structure [6][24][25]. The role of FLG is to generate natural moisturizing factors (NMFs) and to provide a scaffold for the extracellular lipid matrix. If filaggrin is not synthetized correctly or if there is a mutation, then the differentiation of keratinocytes will be dysregulated, and the barrier will be compromised. Not only keratinocytes are involved, but lipid alteration, a reduction in moisturizing agents and barrier disruption are other characteristics present in AD. The consequence of these factors is that the skin is exposed to different infections and pathogens. Moreover, if the SC barrier is not structurally uniform, then keratinocytes are not able to create a compact layer, and for this reason, the epidermis will have an increase in transepidermal water loss (TEWL), while skin hydration and the level of NMFs will be lower [26].
It is important to point out that FLG mutations are neither necessary nor sufficient to cause atopic dermatitis; in fact, 60% of patients do not have this mutation [27].
Not only genetic mutations, but other effects, such as the inflammation process and microbiome imbalance, can also contribute to AD development.
2.2. Inflammation Process
AD is a combination of two pathologies: a skin barrier deficit, such as a filaggrin mutation, and an immune dysregulation. Different hypotheses have been reported; the first suggests that immunological aberrations are the primary phase leading to the development of the disease, with the skin barrier being affected. The second one states that a compromised epidermal barrier primarily leads to the onset of topical eczema and secondarily to immune dysregulation [28].
In particular, AD follows a two-step process: from an acute to a chronic state. In the initial, acute phase, T-helper 2 and T-helper 22 cell responses are increased in the skin, with some involvement of T-helper 17 cells. The mediators produced can influence skin inflammation, and they contribute to the impairment of the skin barrier. Moreover, they activate different cell types, such as keratinocytes, which increase skin inflammation through the release of proinflammatory cytokines. The disease continues its progression in a chronic manner, in which type 1 immunity prevails with Th1 pathways and a still important contribution from T-helper 2 cells [29][30].
2.3. Microbiome in AD
In addition to genetic evidence, the microbiota also plays a key role in the development of skin diseases. In fact, the perturbation of resident flora can determine the colonization of some pathogenic species. A diverse composition of the microbiota can also alter the epidermal barrier.
The bacterium present in the highest percentage in atopic skin is S. aureus. It is able to produce toxins, enzymes and antigens capable of bypassing the immune system and the skin barrier. Its enterotoxins can induce the expansion of T- and B-cells, leading to the production of inflammatory cytokines., e.g., toxins and leucocidins. They initiate the process of cytokine production, hemolysis and leukocyte death. β-toxin, on the other hand, causes cell death, inflammation and destruction of the skin barrier. Researchers can say that these substances help S. aureus to grow and survive by targeting the host’s immune response and the integrity of the skin barrier [31][32][33].
A study conducted by Amy S. Paller et al. confirmed that in AD, an over-colonization of S. aureus (Gram-positive aerobe bacterium) occurs [34]. Thanks to epidemiological, metagenomic and functional studies, it has been shown that the link between this bacterium and AD is sophisticated and depends on host and pathogen factors. While some host factors (physical, antimicrobial) aid in the maintenance of healthy skin, this bacterium can adhere and invade the epidermis, contributing to and promoting the development of an inflammatory state [35].
In an atopic patient, in fact, the percentage of this bacterium is found to be higher, with an increase from 30 to 100% depending on the type of patient, the size of the sample site taken, and the method used for microbial analysis. In the literature, different sampling methods have been observed to analyze the bacterial composition. However, heterogeneous methods have led to different results.
Other results have suggested that when analyzing the skin microbiota with a deep shotgun metagenomic sequencing, the microbial diversity is lower during an AD flare. In particular, in inflamed atopic skin, the genera Streptococcus, Corynebacterium, Cutibacterium and the phylum Proteobacteria decreased with respect to the genus Staphylococcus [35].
Instead, if researchers consider bacterial-culture-based methods, a meta-analysis including 95 studies demonstrated a different distribution of this bacterium on the same patient in injured areas (70%) than in the uninjured areas (39%) [36].
In fact, there is still no real cure that targets the different bacterial composition in AD, and for this reason, possible future therapies could be those in which the precise target is the skin microbiota, with the aim of restoring the commensal species to bring back a healthy and balanced skin environment [37].
Other studies were performed to investigate the microbiota in AD. For example, Kwon et al. [38] evaluated differences in the skin surface microbiome in 18 patients (aged 5–40 years old), prescribing TCS (methylprednisolone cream) and oral histamine. The microbiome was compared between lesioned and non-lesioned skin at different time intervals. The results showed that the proportion of the genus Staphylococcus, which was >80% in baseline lesioned skin, decreased drastically after treatment (week six) and increased slightly after the discontinuation of treatment (week nine). Moreover, the proportion was much higher in lesioned than in non-lesioned skin, even after treatment. In baseline lesioned skin, S. aureus comprised 72.5% of the total species, while S. epidermidis and S. caprae were the second and third most common species, respectively. In non-lesioned skin, Cutibacterium acnes was the most common species, followed by S. aureus and S. epidermidis. The proportion of S. aureus on lesioned and non-lesioned skin was significantly different (p = 0.0014). As a result, researchers can state that this bacterium was more present in lesioned skin at all time points, including week six. The proportion of S. epidermidis, on the other hand, in baseline lesioned and non-lesioned skin was 6.3% and 7.1%, respectively.
This study highlighted how the microbiota changes under injury conditions. In fact, it confirmed that in AD with wounds, S. aureus is the species that predominates with respect to non-lesioned skin, where the composition is different and where Cutibacterium acnes prevails. Moreover, Kwon et al. demonstrated that topical cream, such as TCS, in combination with oral histamine can decrease the colonization of S. aureus.
Previously, researchers stated that different sampling methods can lead to different results. The most common method used is swabs, which allow an analysis of the surface microbiota. Other methods (for example, strips as a non-invasive method, and skin biopsies as a more invasive method) can be used to study the microbial composition in the deeper layers of the epidermis.
In the work of Martin et al. [39], 23 children (six months old with a familiar predisposition) were enrolled. This study aimed to investigate skin parameters and microbiota composition changes with (n = 11) or without (n = 12) the use of an emollient. Parameters were evaluated only after six months. For the skin results, pH and TEWL were measured. In both cases, they were lower in the emollient group than the control one, while the skin water capacitance was higher in the emollient group [38]. The lower value of TEWL should be considered as a positive aspect, as this correlates with the restoration of the skin barrier and the firmness of corneocytes.
The main shortcoming of these studies is related to the duration of treatment. In fact, since the use of the product lasted for only six months, it is not possible to predict the long-term effects [38]. For example, researchers cannot affirm if, with the application of the product, the barrier function will be restored completely. For microbiota analyses, skin samples were obtained by using a flocked swab. Bacterial DNA was amplified and later sequenced, targeting the 16S rRNA gene, in particular regions V1-V3, which allowed the bacterial discriminations. The S. mitis group and S. salivarius were shown to be present in different compositions between samples from the emollient and control groups, while Streptococcus predominated in both the emollient and the control groups.
S. salivarius was significantly higher in the emollient group than in the controls at all sampling sites (cheek, p = 0.02; dorsal forearm, p = 0.02; volar forearm, p = 0.02); instead, S. mitis was less shown in the emollient group than in the controls, without any particular significance [38][39].
The next step was the analysis of S. salivarius in the healthy neonates and in patients with AD in the control group. The proportion of this bacterium in the cheeks appeared lower in AD infants as compared to healthy ones. The decrease in the S. salivarius proportion associated with AD was not statistically significant, presumably due to the low number of infants with AD (n = 3).
Although the limitation of this study has already been mentioned, the hypothesis is that the long-term use of the product with emollient effects, leading to a decrease in pH and an increase in S. salivarius, may help children with atopic dermatitis to avoid the onset of more serious symptoms.
With these studies, it can be considered how different analysis methods and microbiota sampling can lead to different results. In fact, if in the first study researchers focus on Staphylococcus aureus as the main exponent present in a higher percentage of AD patients, then in the last study, Staphylococcus salivarius and mitis are emphasized as the major species. As a result, it can be stated that it is necessary to find a standardized sampling method to analyze the bacterial composition, and thus the skin microbiota at a deeper level, that is, not focusing on species only but also the subspecies and phylotype levels. In this way, researchers can better understand what the real players are that are present on an atypical skin sample, identifying the genetic characteristics.
Treatments for this therapy are complex, and they vary depending on the patient’s condition, age, compliance, and cost. Treatments are also divided into topical and systemic. Firstly, the use of emollients and moisturizers is intended to provide hydration, repair the skin barrier and reduce itching, redness and flaking [40][41][42].
These products usually contain a high percentage of oils. Another recommendation is to take baths once a day for 5 to 10 min in lukewarm water. In more severe cases, dermatologists suggest adding bleach (0.005%) twice a week to counteract the over-colonization of S. aureus, as it has antiseptic properties. If researchers consider medication, the first line of treatment is corticosteroids, which have an anti-inflammatory effect on immune cells such as T-cells and macrophages. Different types of corticosteroids can be used depending on the stage of the disease: in more severe cases, class I corticosteroids are recommended, while in milder cases, patients can take class VII corticosteroids. The latter are often applied in children or in more sensitive areas of the body, where the skin is thinner. These drugs are suggested as they are able to contrast the growth of S. aureus by inhibiting the production of the peptides it produces. Doctors usually recommend brief use because they can lead to side effects such as redness, striae, and atrophy [43][44].
The routes of administration can be different. Indeed, they can be applied directly to atopic skin, or delivered through wet wraps, allowing the active ingredient to penetrate more deeply, at the same time providing protection to compromised skin and lowering transepidermal water loss. In the most extreme cases, topical application is not sufficient. Consequently, the systemic administration of immunomodulators such as cyclosporine and azathioprine is used. The last approach, which could be considered as an adjuvant, is phototherapy. For this purpose, UVB irradiation (NB-UVB/UVB 311 nm) and medium-dose ultraviolet radiation (UVA1) are used [45][46][47].
References
- Ederveen, T.H.A.; Smits, J.P.H.; Boekhorst, J.; Schalkwijk, J.; Van den Bogaard, E.H.; Zeeuwen, P. Skin microbiota in health and disease: From sequencing to biology. J. Dermatol. 2020, 47, 1110–1118.
- Egert, M.; Simmering, R.; Riedel, C.U. The association of the skin microbiota with health, immunity, and disease. Clin. Pharmacol. Ther. 2017, 102, 62–69.
- Chen, Y.E.; Fischbach, M.A.; Belkaid., Y. Skin microbiota-host interactions. Nature 2018, 553, 427–436.
- Balato, A.; Cacciapuoti, S.; Di Caprio, R.; Marasca, C.; Masarà, A.; Raimondo, A.; Fabbrocini, G. Human microbiome: Composition and role in inflammatory skin diseases. Arch. Immunol. Ther. Exp. 2019, 67, 1–18.
- Chen, Y.H.; Tsao, H. The skin microbiome: Current perspectives and future challenges. J. Am. Acad. Dermatol. 2013, 69, 143–155.
- Avena-Woods, C. Overview of atopic dermatitis. Am. J. Manag. Care 2017, 23, 115–123.
- Picardo, M.; Ottaviani, M. Skin microbiome and skin disease. J. Clin. Gastroenterol. 2014, 48, 85–86.
- Musthaq, S.; Mazuy, A.; Jakus, J. The microbiome in dermatology. Clin. Dermatol. 2018, 36, 390–398.
- Baldwin, H.E.; Bhatia, N.D.; Friedman, A.; Eng, R.M.; Seite, S. The Role of Cutaneous Microbiota Harmony in Maintaining a Functional Skin Barrier. J. Drug. Dermatol. 2017, 16, 12–18.
- Belkaid, Y.; Segre, J.A. Dialogue between skin microbiota and immunity. Science 2014, 346, 954–959.
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377.
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologist. J. Eur. Acad. Dermatol. Venerol. 2016, 30, 2038–2047.
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155.
- Clavaud, C.; Jourdain, R.; Bar-Hen, A.; Tichit, M.; Bouchier, C.; Pouradier, F.; El Rawadi, C.; Guillot, J.; Ménard-Szczebara, F.; Breton, L.; et al. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS ONE. 2013, 8, e58203.
- Shibagaki, N.; Suda, W.; Clavaud, C.; Bastien, P.; Takayasu, L.; Iioka, E.; Kurokawa, R.; Yamashita, N.; Hattori, Y.; Shindo, C.; et al. Aging-related changes in the diversity of women’s skin microbiomes associated with oral bacteria. Sci. Rep. 2017, 7, 10567.
- Xu, Z.; Wang, Z.; Yuan, C.; Liu, X.; Yang, F.; Wang, T.; Wang, J.; Manabe, K.; Qin, O.; Wang, X.; et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep. 2016, 6, 24877.
- Zhang, E.; Tanaka, T.; Tajima, M.; Tsuboi, R.; Nishikawa, A.; Sugita, T. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol. Immunol. 2011, 55, 625–632.
- Schommer, N.N.; Gallo, R.L. Structure and function of the human skin microbiome. Trends Microbiol. 2013, 21, 660–668.
- Egert, M.; Simmering, R. The Microbiota of the Human Skin. Adv. Exp. Med. Biol. 2016, 902, 61–81.
- Kim, B.E.; Leung, D.Y.M. Significance of skin barrier dysfunction in atopic dermatitis. Allergy Asthma Immunol. Res. 2018, 10, 207–215.
- Urban, K.; Chu, S.; Giesey, R.L.; Mehrmal, S.; Uppal, P.; Nedley, N.; Delost, G.R. The global, regional, and national burden of atopic dermatitis in 195 countries and territories: An ecological study from the Global Burden of Disease Study. JAAD Int. 2020, 2, 12–18.
- Valdman-Grinshpoun, Y.; Ben-Amitai, D.; Zvulunov, A. Barrier-restoring therapies in atopic dermatitis: Current approaches and future perspectives. Dermatol. Res. Pract. 2012, 923134.
- Brunner, P.M.; Leung, D.Y.M.; Guttman-Yassky, E. Immunologic, microbial, and epithelial interactions in atopic dermatitis. Ann. Allergy Asthma Immunol. 2018, 120, 34–41.
- Lowe, A.J.; Leung, D.Y.M.; Tang, M.L.K.; Su, J.C.; Allen, K.J. The skin as a target for prevention of the atopic march. Ann. Allergy Asthma Immunol. 2018, 120, 145–151.
- Fujii, M. Current understanding of pathophysiological mechanisms of atopic dermatitis: Interactions among skin barrier dysfunction, immune abnormalities and pruritus. Biol. Pharm. Bull. 2020, 43, 12–19.
- Čepelak, I.; Dodig, S.; Pavić, I. Filaggrin and atopic march. Biochem. Med. 2019, 29, 214–227.
- Archer, N.K.; Jo, J.H.; Lee, S.K.; Kim, D.; Smith, B.; Ortines, R.V.; Wang, Y.; Marchitto, M.C.; Ravipati, A.; Cai, S.S.; et al. Injury, dysbiosis, and filaggrin deficiency drive skin inflammation through keratinocyte IL-1α release. J. Allergy Clin. Immunol. 2019, 143, 1426–1443.
- Sroka-Tomaszewska, J.; Trzeciak, M. Molecular Mechanisms of Atopic Dermatitis Pathogenesis. Int. J. Mol. Sci. 2021, 22, 4130.
- Higashi, N.; Gesser, B.; Kawana, S. Expression of IL-18 mRNA and secretion of IL-18 are reduced in monocytes from patients with atopic dermatitis. J. Allergy Clin. Immunol. 2001, 108, 607–614.
- Van Reijsen, F.C.; Bruijnzeel-Koomen, C.A.; Kalthoff, F.S. Skin-derived aeroallergen-specific T-cell clones of Th2 phenotype in patients with atopic dermatitis. J. Allergy Clin. Immunol. 1992, 90, 184–193.
- Kobayashi, T.; Glatz, M.; Horiuchi, K.; Kawasaki, H.; Akiyama, H.; Kaplan, D.H.; Kong, H.H.; Amagai, M.; Nagao, K. Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity 2015, 42, 756–766.
- Salvati, L.; Cosmi, L.; Annunziato, F. From Emollients to Biologicals: Targeting Atopic Dermatitis. Int. J. Mol. Sci. 2021, 22, 10381.
- Gittler, J.K.; Shemer, A.; Suárez-Fariñas, M.; Fuentes-Duculan, J.; Gulewicz, K.J.; Wang, C.Q.; Mitsui, H.; Cardinale, I.; de Guzman Strong, C.; Krueger, J.G.; et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J. Allergy Clin. Immunol. 2012, 130, 1344–1354.
- McLean, W.H.; Irvine, A.D. Heritable Filaggrin Disorders: The Paradigm of Atopic Dermatitis. J. Investig. Dermatol. 2012, 132, 20–21.
- Scharschmidt, T.C.; Man, M.Q.; Hatano, Y.; Crumrine, D.; Gunathilake, R. Filaggrin deficiency confers a paracellular barrier abnormality that reduces inflammatory theresholds to irritants and haptens. J. Allergy Clin. Immunol. 2009, 124, 496–506.
- Nguyen, H.L.T.; Trujillo-Paez, J.V.; Umehara, Y.; Yue, H.; Peng, G.; Kiatsurayanon, C.; Chieosilapatham, P.; Song, P.; Okumura, K.; Ogawa, H.; et al. Role of Antimicrobial Peptides in Skin Barrier Repair in Individuals with Atopic Dermatitis. Int. J. Mol. Sci. 2020, 21, 7607.
- Weidinger, S.; Novak, N. Atopic dermatitis. Lancet 2016, 387, 1109–1122.
- Paller, A.S.; Kong, H.H.; Seed, P.; Naik, S.; Scharschmidt, T.C.; Gallo, R.L.; Luger, T.; Irvine, A.D. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 26–35.
- Geoghegan, J.A.; Irvine, A.D.; Foster, T.J. Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends Microbiol. 2018, 26, 484–497.
- Torres, T.; Ferreira, E.O.; Gonçalo, M.; Mendes-Bastos, P.; Selores, M.; Filipe, P. Update on Atopic Dermatitis. Acta Med. Port. 2019, 32, 606–613.
- Frazier, W.; Bhardwaj, N. Atopic Dermatitis: Diagnosis and Treatment. Am. Fam. Physician. 2020, 101, 590–598.
- González-López, G.; Ceballos-Rodríguez, R.M.; González-López, J.J.; Feito Rodríguez, M.; Herranz-Pinto, P. Efficacy and safety of wet wrap therapy for patients with atopic dermatitis: A systematic review and meta-analysis. Br. J. Dermatol. 2017, 177, 688–695.
- Silverberg, N.B. Atopic dermatitis prevention and treatment. Cutis 2017, 100, 173–177, 192.
- Totté, J.E.; Van Der Feltz, W.T.; Hennekam, M.; van Belkum, A.; van Zuuren, E.J.; Pasmans, S.G. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: A systematic review and meta- analysis. Br. J. Dermatol. 2016, 175, 687–695.
- Huang, J.T.; Abrams, M.; Tlougan, B.; Rademaker, A.; Paller, A.S. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics 2009, 123, e808–e814.
- Kwon, S.; Choi, J.Y.; Shin, J.W.; Huh, C.H.; Park, K.C.; Du, M.H.; Yoon, S.; Na, J.I. Changes in Lesional and Non-lesional Skin Microbiome During Treatment of Atopic Dermatitis. Acta Derm. Venereol. 2019, 3, 284–290.
- Glatz, M.; Jo, J.H.; Kennedy, E.A.; Polley, E.C.; Segre, J.A.; Simpson, E.L.; Kong, H.H. Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLoS ONE. 2018, 13, e0192443.
More
Information
Subjects:
Dermatology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.3K
Revisions:
2 times
(View History)
Update Date:
27 Oct 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




