Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Miguel Alcoceba | -- | 1191 | 2022-10-26 16:21:28 | | | |
| 2 | Conner Chen | + 20 word(s) | 1211 | 2022-10-27 03:45:48 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Alcoceba, M.; García-Álvarez, M.; Okosun, J.; Ferrero, S.; Ladetto, M.; Fitzgibbon, J.; García-Sanz, R. Cell of Origin and Pathogenesis of FL Transformation. Encyclopedia. Available online: https://encyclopedia.pub/entry/31459 (accessed on 07 February 2026).
Alcoceba M, García-Álvarez M, Okosun J, Ferrero S, Ladetto M, Fitzgibbon J, et al. Cell of Origin and Pathogenesis of FL Transformation. Encyclopedia. Available at: https://encyclopedia.pub/entry/31459. Accessed February 07, 2026.
Alcoceba, Miguel, María García-Álvarez, Jessica Okosun, Simone Ferrero, Marco Ladetto, Jude Fitzgibbon, Ramón García-Sanz. "Cell of Origin and Pathogenesis of FL Transformation" Encyclopedia, https://encyclopedia.pub/entry/31459 (accessed February 07, 2026).
Alcoceba, M., García-Álvarez, M., Okosun, J., Ferrero, S., Ladetto, M., Fitzgibbon, J., & García-Sanz, R. (2022, October 26). Cell of Origin and Pathogenesis of FL Transformation. In Encyclopedia. https://encyclopedia.pub/entry/31459
Alcoceba, Miguel, et al. "Cell of Origin and Pathogenesis of FL Transformation." Encyclopedia. Web. 26 October, 2022.
Copy Citation
The gold standard for determining follicular lymphoma (FL) transformation is based on the histologically confirmed progression of grade 1, 2, or 3A FL to a high-grade lymphoma, consisting of a predominance of large cells and the loss of the follicular architecture. Histological transformation (HT) to a more aggressive disease–mostly diffuse large B-cell lymphoma–is considered one of the most dismal events in the clinical course of FL.
transformed follicular lymphoma
genetics
histological transformation
1. Definition of FL Transformation
The gold standard for determining follicular lymphoma (FL) transformation is based on the histologically confirmed progression of grade 1, 2, or 3A FL to a high-grade lymphoma, consisting of a predominance of large cells and the loss of the follicular architecture [1][2]. Most of the transformed cases have a diffuse large B-cell lymphoma (DLBCL) histology (>80% of the cases) according to the current WHO classification, although other histologies have been described, such as high-grade B-cell lymphoma, FL grade 3B, Burkitt lymphoma, B lymphoblastic leukemia/lymphoma, and plasmablastic lymphoma [2][3][4][5]. There are other atypical forms suggestive of histological transformation, such as the presence at diagnosis of both FL and DLBCL cells, at the same site, referred to as composite lymphoma, or at different sites such as DLBCL in the lymph node and FL in bone marrow, as well as DLBCL cases that undergo a process of reverse transformation, relapsing as a lower grade lymphoma. These forms are not addressed in these contents.
Since lymphoma lesions are not isolated, other tumour areas might have a FL component at the same time in addition to the transformation area [1][6][7]. Positron emission tomography and computerized tomography (PET/CT) could help by selecting the biopsy site according to the highest standardized uptake value (SUV) of 18[F] fluorodeoxyglucose, since a high value (generally > 14) is correlated with more aggressive histology [8]. However, only ~50% of patients are biopsied, with inaccessibility of the tumour, the patient’s clinical situation or refusal among the main reasons [9]. Based on the clinical behaviour of transformed patients, several clinical criteria of transformation suspicion could be of utility in these cases, including an increase in lactate dehydrogenase (LDH) levels or hypercalcemia, rapid lymphadenopathy growth or the appearance of lymphoma masses or conglomerates, and the novel involvement of extranodal sites and new B symptoms. However, these criteria vary between studies and are not standardised [10][11][12]. Moreover, these clinical criteria are also present in patients who progress without transformation [9].
2. Cell of Origin and Pathogenesis of FL Transformation
Transformed cases may have changes in their immunophenotype, with an antigenic drift including CD10 loss or positivity of MUM1/IRF4. Although most transformed FLs are of germinal B-cell DLBCL subtype (GCB), up to 15–20% of the cases change to an activated B-cell (ABC) without differences in survival between both subtypes [4][13][14]. This contrasts with transformation in other B-cell lymphoproliferative disorders, such as chronic lymphocytic leukaemia (CLL), Waldenström macroglobulinemia or marginal zone lymphoma, in which transformed cases are mostly ABC/non-GCB [15][16][17].
Recurrent rearranged genes in DLBCL include BCL2, BCL6 and MYC. There are no major changes in the frequency of BCL2 or BCL6 translocations in transformed samples compared to FL diagnosis, however, MYC translocations are commonly acquired and are present in 25% of transformed cases [4][18]. The acquisition of MYC translocations implies an increase in the proportion of double-hit lymphomas (presence of both BCL2 and MYC translocations) in transformed patients, which is associated with a shorter survival from transformation (SFT) [4], although differences were not statistically significant likely due to the low number of cases analysed.
High-resolution genome wide analysis using SNP-array, whole-genome (WGS) or whole-exome sequencing (WES), and targeted next-generation sequencing studies in transformed FL identify increased genomic complexity and mutational burden at transformation in comparison to FL [5][18][19][20][21][22][23][24]. The most recurrent genetic lesions acquired in transformed FL cases are summarized in Table 1 and Figure 1, and include alterations (mainly mutations and/or deletions) in TP53 in approximately 15–30% transformed cases, CDKN2A/B deletions in 20–30% of cases, and B2M mutations and/or deletions in 20–25% of cases, together with the previously mentioned MYC translocations [5][18][19][20][22][23][25]. Of note, although these lesions are commonly acquired in transformation, they are not specific, as they could also be present at diagnosis or acquired during disease recurrence, representing markers of more aggressive disease [2][5][18][19][20][22][23][26]. In fact, these are also common acquired lesions in refractoriness and/or transformation in other haematological disorders [27][28].
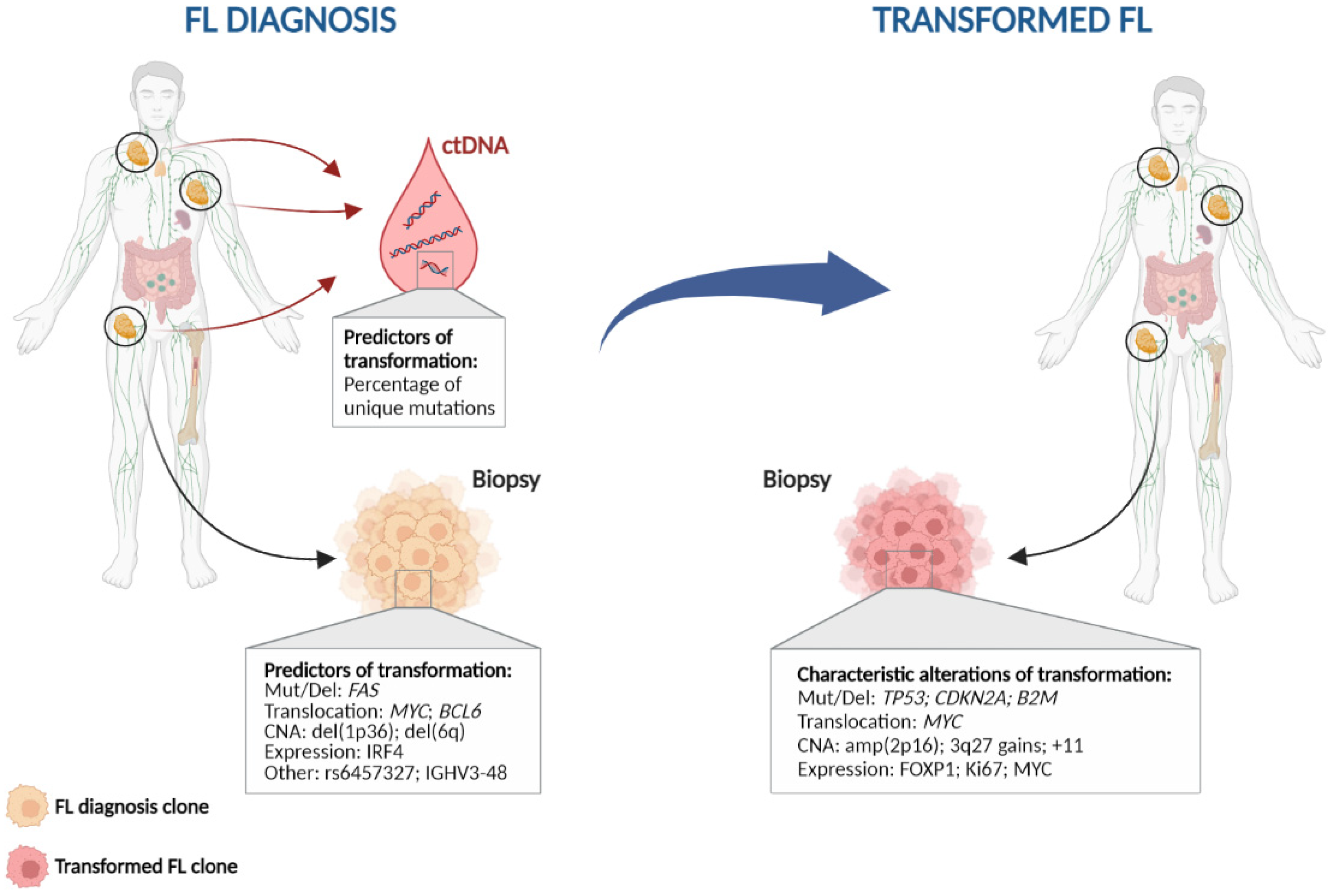
Figure 1. Characteristic genetic events in follicular lymphoma histological transformation. Left: Potential predictors of transformation; Right: Genetic alterations commonly found at histological transformation. CNA: copy number alteration; ctDNA: circulating tumour DNA; Mut/Del: mutation and/or deletion.
Other commonly acquired events include mutations in MYC, CCND3, CD58, EBF1, GNA13, P2RY8, and S1PR2, as well as gains of 3q27.3-q28 (BCL6), amplification of 2p16 (REL), and gains in chromosomes 2, 5, and 11 [5][18][20][22][23][29][30][31]. All of these alterations together indicate that different pathways may be involved in transformation, including both cell cycle and DNA damage dysregulation, immune escape, JAK-STAT or NF-κB pathways, increased proliferation, and lymphoma cell migration.
When transformed cases are classified according to their cell-of-origin, different patterns of mutations are observed in each group. MYD88, CD79B, and BCL10 mutations are more frequently (~15–25%) identified in ABC transformed cases, while amplification of 2p16 (REL) are more common in GCB, consistent with what is observed in DLBCL [5][22][23][31]. This suggests that there could be at least two different subgroups of transformed FLs. Moreover, recent studies have classified de novo DLBCL into different molecular clusters according to their mutation, copy-number and structural variation profile, and these clusters are associated with different outcomes [32][33]. There is no information regarding the distribution of these clusters in transformed FL, although some of the most common alterations in histological transformation (HT) such as TP53 mutations/deletions, CDKN2A/B deletions and REL amplification are present in cluster C2, while C5 and MCD comprised mostly ABC-DLBCL, with mutations in CD79B and MYD88 [32][33]. This suggests that different clusters of transformed FL could be present, possibly with different pathways leading to transformation, and perhaps a distinct outcome. In line with this, a previous study showed an increased proliferation rate by gene expression analysis at transformation in a subgroup of HT, which was enriched with aberrations in TP53, CDKN2A/B, and REL in contrast to other HT, suggesting different mechanisms of transformation [13].
Table 1. Biological and genetic factors enriched at follicular lymphoma histological transformation in the literature.
| Category | Variable | Biological Effect | Effect on Transformation |
|---|---|---|---|
| IHQ and microenvironment | IRF4 expression | - | Increased at HT [4] |
| MYC expression | - | Increased at HT [34] | |
| FOXP1 expression | - | Increased at HT [35] | |
| Genomic variants | TP53 mutation and deletion | Cell cycle | Increased at HT [5][18][19][20][23] |
| B2microglobulin mutation and deletion | Immune surveillance | Increased at HT [5][18] | |
| FAS mutation and deletion | Apoptosis | Enriched in transformed cases [18] | |
| MYC mutation and translocation | Cell cycle | Increased at HT [4][18] | |
| CCND3 mutation | Cell cycle, JAK-STAT signalling | Increased at HT [5][19] | |
| EBF1 mutation | B-cell development | Increased at HT [5][20] | |
| GNA13 mutation | NF-kB/BCR signalling | Increased at HT [5] | |
| P2RY8 mutation | B-cell migration | Increased at HT [5] | |
| S1PR2 mutation | Proliferation | Increased at HT [5] | |
| CD58 mutation | Immune surveillance | Increased at HT [18] | |
| MYD88 mutation | NF-kB/BCR signalling | Increased at HT, ABC-HT related [5][19][20][23] | |
| CD79B mutation | NF-kB/BCR signalling | Increased at HT, ABC-HT related [5][19][23] | |
| BCL10 mutation | NF-kB/BCR signalling | Increased at HT, ABC-HT related [5][23] | |
| CDKN2A/B deletion | Cell cycle | Increased at HT [5][18][20][22][25][31] | |
| BCL6 translocation | B-cell differentiation | Increased at HT [4][36] | |
| 2p16 (REL) amplification | NF-kB/BCR signalling | Increased at HT, GCB-HT related [18][22][29][30][31] | |
| 3q27.3-q28 (BCL6) gains | B-cell differentiation | Increased at HT [18][22][29][30] | |
| Chromosomes 2, 5 and 11 gains | - | Increased at HT [22][29] | |
| Genomic complexity -copy-number changes- | - | Increased at HT [5][18][20][21][22][23][24] | |
| Genetic complexity -mutations- | - | Increased at HT [5][18][19][20][23][37] |
HT: Histological transformation.
References
- Casulo, C.; Burack, W.R.; Friedberg, J.W. Transformed follicular non-Hodgkin lymphoma. Blood 2015, 125, 40–47.
- Lossos, I.S.; Gascoyne, R.D. Transformation of follicular lymphoma. Best Pract. Res. Clin. Haematol. 2011, 24, 147–163.
- Ouansafi, I.; He, B.; Fraser, C.; Nie, K.; Mathew, S.; Bhanji, R.; Hoda, R.; Arabadjief, M.; Knowles, D.; Cerutti, A.; et al. Transformation of follicular lymphoma to plasmablastic lymphoma with c-myc gene rearrangement. Am. J. Clin. Pathol. 2010, 134, 972–981.
- Kridel, R.; Mottok, A.; Farinha, P.; Ben-Neriah, S.; Ennishi, D.; Zheng, Y.; Chavez, E.A.; Shulha, H.P.; Tan, K.; Chan, F.C.; et al. Cell of origin of transformed follicular lymphoma. Blood 2015, 126, 2118–2127.
- Kridel, R.; Chan, F.C.; Mottok, A.; Boyle, M.; Farinha, P.; Tan, K.; Meissner, B.; Bashashati, A.; McPherson, A.; Roth, A.; et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016, 13, e1002197.
- Salles, G.; Coiffier, B. Histologic transformation in follicular lymphoma. Ann. Oncol. 1998, 9, 803–805.
- Araf, S.; Wang, J.; Korfi, K.; Pangault, C.; Kotsiou, E.; Rio-Machin, A.; Rahim, T.; Heward, J.; Clear, A.; Iqbal, S.; et al. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia 2018, 32, 1261–1265.
- Bodet-Milin, C.; Kraeber-Bodere, F.; Moreau, P.; Campion, L.; Dupas, B.; Le, G.S. Investigation of FDG-PET/CT imaging to guide biopsies in the detection of histological transformation of indolent lymphoma. Haematologica 2008, 93, 471–472.
- Gine, E.; Montoto, S.; Bosch, F.; Arenillas, L.; Mercadal, S.; Villamor, N.; Martinez, A.; Colomo, L.; Campo, E.; Montserrat, E.; et al. The Follicular Lymphoma International Prognostic Index (FLIPI) and the histological subtype are the most important factors to predict histological transformation in follicular lymphoma. Ann. Oncol. 2006, 17, 1539–1545.
- Bastion, Y.; Sebban, C.; Berger, F.; Felman, P.; Salles, G.; Dumontet, C.; Bryon, P.A.; Coiffier, B. Incidence, predictive factors, and outcome of lymphoma transformation in follicular lymphoma patients. J. Clin. Oncol. 1997, 15, 1587–1594.
- Al-Tourah, A.J.; Gill, K.K.; Chhanabhai, M.; Hoskins, P.J.; Klasa, R.J.; Savage, K.J.; Sehn, L.H.; Shenkier, T.N.; Gascoyne, R.D.; Connors, J.M. Population-based analysis of incidence and outcome of transformed non-Hodgkin’s lymphoma. J. Clin. Oncol. 2008, 26, 5165–5169.
- Link, B.K.; Maurer, M.J.; Nowakowski, G.S.; Ansell, S.M.; Macon, W.R.; Syrbu, S.I.; Slager, S.L.; Thompson, C.A.; Inwards, D.J.; Johnston, P.B.; et al. Rates and outcomes of follicular lymphoma transformation in the immunochemotherapy era: A report from the University of Iowa/MayoClinic Specialized Program of Research Excellence Molecular Epidemiology Resource. J. Clin. Oncol. 2013, 31, 3272–3278.
- Davies, A.J.; Rosenwald, A.; Wright, G.; Lee, A.; Last, K.W.; Weisenburger, D.D.; Chan, W.C.; Delabie, J.; Braziel, R.M.; Campo, E.; et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma proceeds by distinct oncogenic mechanisms. Br. J. Haematol. 2007, 136, 286–293.
- Maeshima, A.M.; Taniguchi, H.; Toyoda, K.; Yamauchi, N.; Makita, S.; Fukuhara, S.; Munakata, W.; Maruyama, D.; Kobayashi, Y.; Tobinai, K. Clinicopathological features of histological transformation from extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue to diffuse large B-cell lymphoma: An analysis of 467 patients. Br. J. Haematol. 2016, 174, 923–931.
- Zanwar, S.; Abeykoon, J.P.; Durot, E.; King, R.; Perez Burbano, G.E.; Kumar, S.; Gertz, M.A.; Quinquenel, A.; Delmer, A.; Gonsalves, W.; et al. Impact of MYD88(L265P) mutation status on histological transformation of Waldenström Macroglobulinemia. Am. J. Hematol. 2020, 95, 274–281.
- Abrisqueta, P.; Delgado, J.; Alcoceba, M.; Oliveira, A.C.; Loscertales, J.; Hernández-Rivas, J.A.; Ferrà, C.; Cordoba, R.; Yáñez, L.; Medina, A.; et al. Clinical outcome and prognostic factors of patients with Richter syndrome: Real-world study of the Spanish Chronic Lymphocytic Leukemia Study Group (GELLC). Br. J. Haematol. 2020, 190, 854–863.
- Bastidas-Mora, G.; Beà, S.; Navarro, A.; Gine, E.; Costa, D.; Delgado, J.; Baumann, T.; Magnano, L.; Rivas-Delgado, A.; Villamor, N.; et al. Clinico-biological features and outcome of patients with splenic marginal zone lymphoma with histological transformation. Br. J. Haematol. 2022, 196, 146–155.
- Pasqualucci, L.; Khiabanian, H.; Fangazio, M.; Vasishtha, M.; Messina, M.; Holmes, A.B.; Ouillette, P.; Trifonov, V.; Rossi, D.; Tabbo, F.; et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014, 6, 130–140.
- García-Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, M.I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Antón, A.; Maldonado, R.; et al. Molecular study of the clonal evolution of follicular lymphoma to aggressive lymphoma. A single center experience. Haematologica 2018, 103, 15.
- Okosun, J.; Bodor, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181.
- O’Shea, D.; O’Riain, C.; Gupta, M.; Waters, R.; Yang, Y.; Wrench, D.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. Regions of acquired uniparental disomy at diagnosis of follicular lymphoma are associated with both overall survival and risk of transformation. Blood 2009, 113, 2298–2301.
- Bouska, A.; McKeithan, T.W.; Deffenbacher, K.E.; Lachel, C.; Wright, G.W.; Iqbal, J.; Smith, L.M.; Zhang, W.; Kucuk, C.; Rinaldi, A.; et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood 2014, 123, 1681–1690.
- Bouska, A.; Zhang, W.; Gong, Q.; Iqbal, J.; Scuto, A.; Vose, J.; Ludvigsen, M.; Fu, K.; Weisenburger, D.D.; Greiner, T.C.; et al. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia 2017, 31, 83–91.
- Qu, X.; Li, H.; Braziel, R.M.; Passerini, V.; Rimsza, L.M.; Hsi, E.D.; Leonard, J.P.; Smith, S.M.; Kridel, R.; Press, O.; et al. Genomic alterations important for the prognosis in patients with follicular lymphoma treated in SWOG study S0016. Blood 2019, 133, 81–93.
- Elenitoba-Johnson, K.S.; Gascoyne, R.D.; Lim, M.S.; Chhanabai, M.; Jaffe, E.S.; Raffeld, M. Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood 1998, 91, 4677–4685.
- Alhejaily, A.; Day, A.G.; Feilotter, H.E.; Baetz, T.; Lebrun, D.P. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin. Cancer Res. 2014, 20, 1676–1686.
- Fabbri, G.; Khiabanian, H.; Holmes, A.B.; Wang, J.; Messina, M.; Mullighan, C.G.; Pasqualucci, L.; Rabadan, R.; Dalla-Favera, R. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J. Exp. Med. 2013, 210, 2273–2288.
- Martello, M.; Poletti, A.; Borsi, E.; Solli, V.; Dozza, L.; Barbato, S.; Zamagni, E.; Tacchetti, P.; Pantani, L.; Mancuso, K.; et al. Clonal and subclonal TP53 molecular impairment is associated with prognosis and progression in multiple myeloma. Blood Cancer J. 2022, 12, 15.
- Eide, M.B.; Liestol, K.; Lingjaerde, O.C.; Hystad, M.E.; Kresse, S.H.; Meza-Zepeda, L.; Myklebost, O.; Troen, G.; Aamot, H.V.; Holte, H.; et al. Genomic alterations reveal potential for higher grade transformation in follicular lymphoma and confirm parallel evolution of tumor cell clones. Blood 2010, 116, 1489–1497.
- Martinez-Climent, J.A.; Alizadeh, A.A.; Segraves, R.; Blesa, D.; Rubio-Moscardo, F.; Albertson, D.G.; Garcia-Conde, J.; Dyer, M.J.; Levy, R.; Pinkel, D.; et al. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood 2003, 101, 3109–3117.
- Kwiecinska, A.; Ichimura, K.; Berglund, M.; Dinets, A.; Sulaiman, L.; Collins, V.P.; Larsson, C.; Porwit, A.; Lagercrantz, S.B. Amplification of 2p as a genomic marker for transformation in lymphoma. Genes Chromosom. Cancer 2014, 53, 750–768.
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690.
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407.
- Aukema, S.M.; van Pel, R.; Nagel, I.; Bens, S.; Siebert, R.; Rosati, S.; van den Berg, E.; Bosga-Bouwer, A.G.; Kibbelaar, R.E.; Hoogendoorn, M.; et al. MYC expression and translocation analyses in low-grade and transformed follicular lymphoma. Histopathology 2017, 71, 960–971.
- Musilova, K.; Devan, J.; Cerna, K.; Seda, V.; Pavlasova, G.; Sharma, S.; Oppelt, J.; Pytlik, R.; Prochazka, V.; Prouzova, Z.; et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood 2018, 132, 2389–2400.
- Akasaka, T.; Lossos, I.S.; Levy, R. BCL6 gene translocation in follicular lymphoma: A harbinger of eventual transformation to diffuse aggressive lymphoma. Blood 2003, 102, 1443–1448.
- García Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Maldonado, R.; Antón, A.; et al. Genetic complexity impacts the clinical outcome of follicular lymphoma patients. Blood Cancer J. 2021, 11, 11.
More
Information
Subjects:
Anatomy & Morphology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
937
Revisions:
2 times
(View History)
Update Date:
27 Oct 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




