Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | José M. Frade | -- | 3824 | 2022-10-24 11:50:50 | | | |
| 2 | Rita Xu | Meta information modification | 3824 | 2022-10-24 12:00:28 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ramón-Landreau, M.; Sánchez-Puelles, C.; López-Sánchez, N.; Lozano-Ureña, A.; Llabrés-Mas, A.M.; Frade, J.M. E2F4DN Transgenic Mice. Encyclopedia. Available online: https://encyclopedia.pub/entry/30955 (accessed on 08 February 2026).
Ramón-Landreau M, Sánchez-Puelles C, López-Sánchez N, Lozano-Ureña A, Llabrés-Mas AM, Frade JM. E2F4DN Transgenic Mice. Encyclopedia. Available at: https://encyclopedia.pub/entry/30955. Accessed February 08, 2026.
Ramón-Landreau, Morgan, Cristina Sánchez-Puelles, Noelia López-Sánchez, Anna Lozano-Ureña, Aina M. Llabrés-Mas, José M. Frade. "E2F4DN Transgenic Mice" Encyclopedia, https://encyclopedia.pub/entry/30955 (accessed February 08, 2026).
Ramón-Landreau, M., Sánchez-Puelles, C., López-Sánchez, N., Lozano-Ureña, A., Llabrés-Mas, A.M., & Frade, J.M. (2022, October 24). E2F4DN Transgenic Mice. In Encyclopedia. https://encyclopedia.pub/entry/30955
Ramón-Landreau, Morgan, et al. "E2F4DN Transgenic Mice." Encyclopedia. Web. 24 October, 2022.
Copy Citation
E2F4 was initially described as a transcription factor with a key function in the regulation of cell quiescence. The regulation of E2F4 is complex, as it can be chemically modified through acetylation, from which researchers present evidence in the brain, as well as methylation, and phosphorylation. The phosphorylation of E2F4 within a conserved threonine motif induces cell cycle re-entry in neurons, while a dominant negative form of E2F4 (E2F4DN), in which the conserved threonines have been substituted by alanines, has been shown to act as a multifactorial therapeutic agent for Alzheimer’s disease (AD).
acetylated E2F4
synapsis
tissue homeostasis
Alzheimer’s disease
5xFAD mice
1. Introduction
E2 factor 4 (E2F4) is a member of the E2F family of transcription factors [1], which are primarily known to regulate the cell cycle. E2F4 was first described as a cell cycle repressor able to interact with p107 [2][3] and p130 [4], two members of the retinoblastoma (RB) family. Nevertheless, its capacity to repress cell cycle progression can be modulated, as it can also facilitate the cell cycle progression of cardiomyocytes, normal intestinal crypt cells, and colorectal cancer cells [5][6]. A number of reviews have been published describing the role of this transcription factor in quiescence and other cell cycle-related mechanisms [7][8][9], and researchers refer to the reader to these informative reviews for this aspect of E2F4 function.
Interestingly, E2F4 can also play other important roles in cellular physiology, including cell and tissue homeostasis and tissue regeneration [7][8][10][11][12]. Therefore, E2F4 can be considered a multifactorial factor with an important impact on neuronal welfare and brain homeostasis [11][12], suggesting that it may be a promising candidate target for neurodegenerative diseases and brain aging.
E2F4 is a phosphoprotein whose phosphorylation within an evolutionary-conserved threonine motif containing T248 (Figure 1) can modify its function [11][13]. This covalent modification has been targeted by substituting T248 and T250 with alanines, thus resulting in a dominant negative form of E2F4 (E2F4DN). This mutant form, or E2F4, prevents cell cycle re-entry in developing neurons [13] and is able to prevent Alzheimer’s disease (AD)-deleterious processes in 5xFAD mice [11], a murine model of this disease [14].
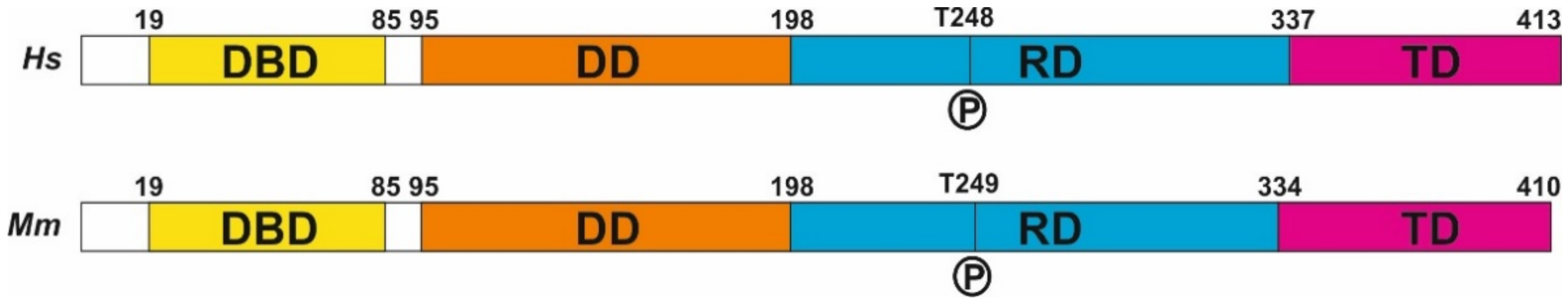
Figure 1. E2F4 structure. The structure of human (Hs) and mouse (Mm) E2F4, derived from NCBI Reference Sequences NP_001941.2 (H. sapiens) and NP_683754.1 (M. musculus). DBD: DNA binding domain, DD: dimerization domain, RD: regulatory domain, TD: transactivation domain.
2. Transcriptional and Non-Transcriptional Functions of E2F4
2.1. E2F4 as a Transcriptional Regulator
Human E2F4 contains 413 amino acids (410 in mouse) distributed throughout four domains (Figure 1). As with other E2F members, it forms a heterodimer with dimerization partner (DP) proteins through its dimerization domain (DD), located at the N-terminus of the molecule. The DD domain is required for its interaction with DNA through the DNA-binding domain (DBD). A third domain located at the C-terminus is required for the function of E2F4 as a transcription factor [15]; this transactivation domain (TD) is blocked when the retinoblastoma (RB) family proteins p107 or p130 interact with E2F4 through its protein-binding domain [10]. This interaction is crucial for the control of the E2F4 function as a transcription factor. Finally, E2F4 has a region that has been proposed as a regulatory domain (RD) [13] in which phosphorylatable residues, such as T248 (see below), are placed. In addition, two nuclear export signals (NES) are present in E2F4, one located within the DBD and the other in the DD [16][17]. These sequences maintain E2F4 within the cytoplasm unless it interacts with p107 or p130, which are required for the translocation of E2F4 to the nucleus. In addition, other factors can induce the translocation of E2F4 to the nucleus, as the latter can also regulate transcription through RB-independent mechanisms [18]. In this regard, E2F4 can interact with KPNB1, RanGAP1, and RanBP2 [18], three proteins that are involved in nuclear import [19][20], and may facilitate E2F4 nuclear translocation in the absence of RB family members. Moreover, E2F4 may be translocated to the nucleus with the help of DP family members DP-2 and DP-3 [21][22][23], likely due to the presence of a nuclear localization signal (NLS) in their sequence, as has been shown in DP-2 [24]. Finally, E2F4 harbors a weak putative NLS in amino acids 52-61 [24], suggesting that E2F4 can translocate into the nucleus in a cofactor-independent manner, similar to E2F5 during keratinocyte differentiation [25].
A ChIP-seq analysis performed in human lymphoblastoid cells identified around 16,000 E2F4 binding sites that potentially regulate 7346 target genes with wide-ranging functions, including cell cycle regulation, DNA repair, RNA processing, stress response, apoptosis, ubiquitination, protein transport and targeting, protein folding, and I-κB kinase/NF-κB cascade regulation [26]. In these cells, E2F4 can bind near transcription start sites (TSSs), a finding confirmed by others [27]. In addition, functional distal sites for E2F4 can be located more than 20 kb away from the annotated TSSs. In both cases, E2F4 can act as an activator as well as a repressor [26]. This analysis also indicated that E2F4 can bind to the promoters of 780 transcription factors, suggesting that E2F4 can indirectly regulate broad classes of genes [26]. Other authors have confirmed that E2F4 can bind to genes related to DNA repair, DNA damage, and G2/M checkpoints, as well as to other classical functions, such as cell cycle regulation, DNA replication, chromosome transactions, and mitotic regulation [28]. In most cases, E2F4 can bind to a specific promoter together with other members of the E2F family [27], indicating that the E2F4 function is subjected to complex cross-regulatory networks [29][30]. Many E2F4 binding sites have been analyzed in specific gene regulatory regions [31]. For instance, the release of a p130-E2F4 complex from sequences immediately upstream of the transcription initiation site of the human CDC2 promoter has been shown to coincide with the induction of CDC2 expression [32].
Several lines of evidence indicate that E2F4 is able to control complex transcriptional regulatory networks in specific cells, thus supporting its multifactorial capacity as a transcription factor. For instance, a combined analysis using gene ontology and expression data has been used to define the network controlled by E2F4 in B cells [33]. In addition, loss-of-function studies on E2F4 silencing using a specific shRNA in acute myeloid leukemia cells have revealed that 276 genes show altered expression patterns in these cells [34]. These E2F4-regulated genes are mostly involved in the regulation of the mitogen-activated protein kinase (MAPK) signaling pathway.
The regulation of gene transcription by E2F4 seems to be mediated through histone acetylation, as E2F4 may interact with CREB binding protein (histone acetyltransferase) [18], and sites where E2F4 binds are histone-modified [26].
2.2. E2F4 and Non-Transcriptional Interactors
E2F4 lacks a strong NLS, which suggests that this protein could play a significant role in the cytoplasm [35]. This is, for instance, the case of the regulation of centriole amplification during multiciliogenesis, which is mediated by the interaction of E2F4 with Deup1 and SAS6, two components of the centriole replication machinery [36]. Indeed, cytoplasmic E2F4 forms organizing centers in multiciliated cells [37]. While centrioles are known to undergo one round of duplication per cell cycle in normal proliferating cells, multiciliated cells show a massive assembly of these organelles during G0, a process initiated by Multicilin in combination with E2F4 (or E2F5) and Dp1 [38][39][40].
The capacity of E2F4 to function out of the nucleus is consistent with a study by Hsu and collaborators [18]. These authors identified a number of E2F4 interactors in mouse embryonic stem cells (mESCs) and a retinal pigment epithelium (RPE)-derived cell line of human origin [18]. Several of these interactors are located outside of the cell nucleus since a cellular component (CC) ontology analysis performed by us using the E2F4 interactors described by Hsu and collaborators [18] confirmed that E2F4 may be functional in the cytoplasm of mESCs and both cytoplasm and extracellular vesicles from RPE-derived cells.
3. Regulation of E2F4 Function by Chemical Modifications
Proteins can be posttranslationally modified through covalent processing events that change their properties, either by proteolytic cleavage or by adding a modifying group, such as acetyl, phosphoryl, glycosyl, and methyl, to one or more amino acids [41]. More than 400 different types of posttranslational modifications [42] affect many aspects of protein function. Some of these chemical modifications have been described in E2F4.
As in the case of other regulators of the cell cycle, E2F4 can be ubiquitinated as a mechanism regulating its protein levels [43]. In addition, E2F4 activity could be modulated by protein acetylation, as observed with another member of the E2F family of transcription factors, E2F1 [44]. E2F1 can be acetylated in sites that lie adjacent to the DBD, thus increasing its DNA-binding ability and activation potential, as well as its protein half-life [44]. In the case of E2F4, Hsu and collaborators [18] demonstrated that both human and mouse E2F4 can be significantly acetylated in K37 and K96. These residues are located within the DBD and DD, respectively, thus suggesting that the capacity for DNA binding and DP heterodimerization of E2F4 can be compromised. This may facilitate the cytoplasmic function of E2F4 as a multifactorial protein. These authors also found small levels of acetylation in K20, K28, K44, K73, K82, K101, K177, K197, K230, and K347 from human E2F4 and in K28, K44, K101, K118, K177, K178, and K339 from mouse E2F4. Most of these residues are located within the DBD and DD of E2F4, suggesting that their acetylation can also participate in the regulation of DNA binding and the DP heterodimerization of E2F4.
Using an acetylated K96-specific antibody, researchers verified that K96 becomes acetylated in some structures of the adult mouse brain in vivo (Figure 2). This form of acetylated E2F4 can be detected in NeuN-positive cells (i.e., neurons) within the hippocampus (dentate gyrus) (Figure 2a), cerebellum (Figure 2b), and NeuN-negative cells located in the rostral migratory stream (RMS) (Figure 2c), likely neural progenitors. Some NeuN-negative cells in the cerebellum also showed acetylated E2F4-specific immunoreactivity (Figure 2b).
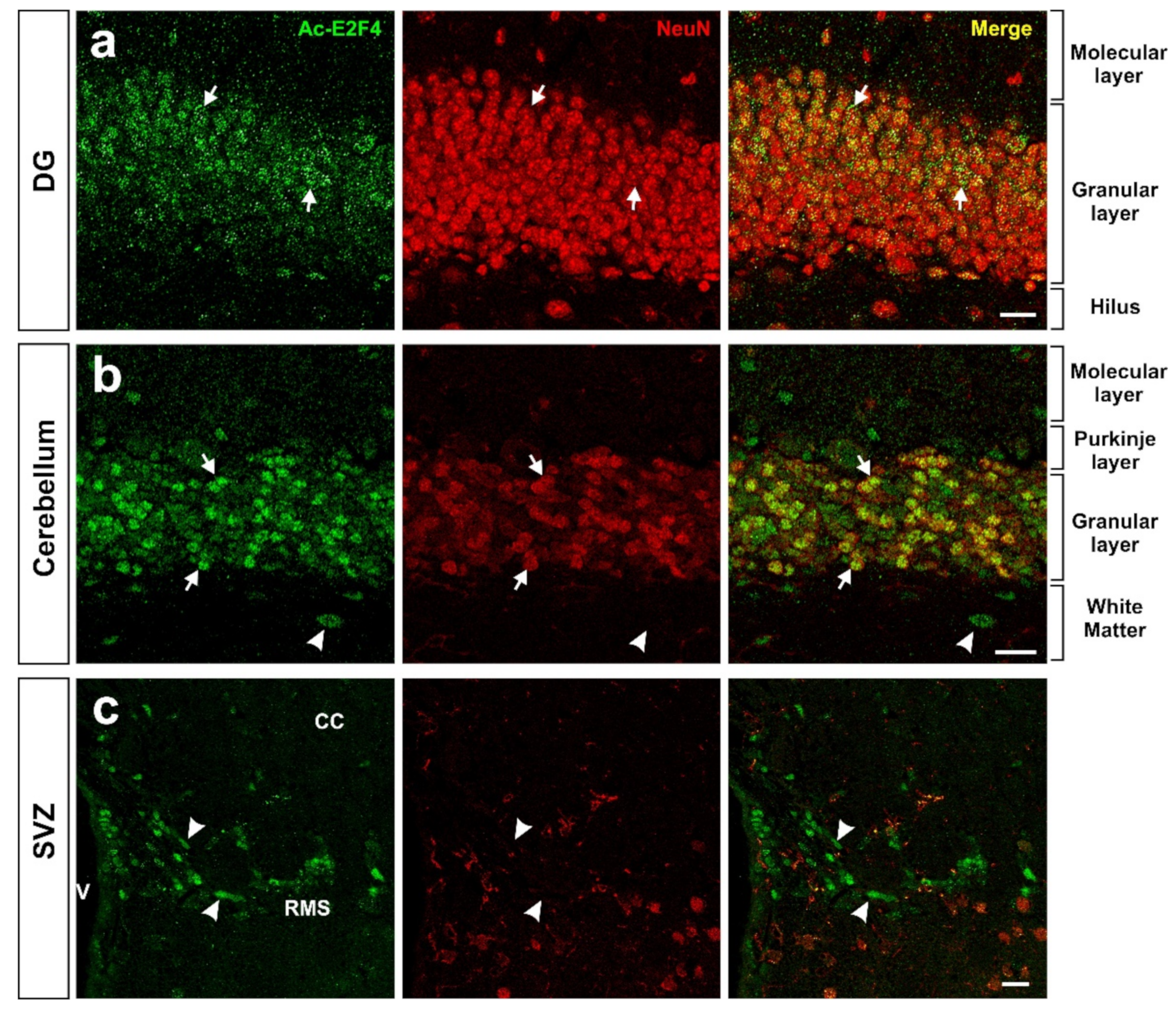
Figure 2. Expression pattern of acetylated E2F4 in the dentate gyrus (DG) (a), cerebellum (b), and subventricular zone (SVZ) (c) of 2.5-month-old WT mice. One single confocal plane showing co-immunostaining with anti-acetylated E2F4 (Ac-E2F4) and anti-NeuN (NeuN) antibodies in sections from the indicated brain areas. NeuN specifically labels neurons. Ac-E2F4 immunostaining in NeuN-positive (arrows) and NeuN-negative (arrowheads) cells is shown. V: ventricle; CC: corpus callosum; RMS: rostral migratory stream. Scale bar: 20 μm.
In non-histone proteins, methylation represents a chemical modification participating in diverse processes, such as cell cycle control, DNA repair, senescence, differentiation, apoptosis, and tumorigenesis [45]. As a multifactorial factor, E2F4 can also become methylated. In this regard, Hsu and collaborators [18] have shown that a significant proportion of K73, K197, and R357 (R360 in mice) residues from E2F4 can be methylated. Interestingly, the methylation of K197 in E2F4 is reminiscent of a similar process in E2F1, affecting K185, which is involved in the regulation of E2F1-induced cell death [45][46][47]. Other residues of human (K20, K37, K53, K57, K74, K96, K101, R147, K177, K230, and K347) and mouse (R297 and K339) E2F4 can also be methylated, as reported by Hsu and collaborators [18].
Finally, the most prominent mechanism regulating E2F4 activity is protein phosphorylation. E2F4 has several residues susceptible to phosphorylation (Figure 1), and several lines of evidence indicate that E2F4 can undergo phosphorylation [48] to modulate its function. In this regard, this chemical modification may regulate E2F4-mediated transcription, either by disrupting its DNA-binding ability, as observed in 3T3 cells [49], or by enhancing the DNA binding of the E2F4/p130 repressor complex, as demonstrated in human fibroblasts [50]. Seven of the theoretical phosphorylation sites of E2F4, including T14, S202, S218, T224, S244, T248, and S384, have been demonstrated to become phosphorylated [51]. Other authors have confirmed the phosphorylation of T14, S218, S244, T248, and S381 in human E2F4 [18], of S218, T224, T249, and S384 in mouse E2F4 [18], and the ortologue of T248/T250 (T261/T263) in chicken E2F4 [13]. In addition, phosphorylation of E2F4 in T249 has been observed in mouse brain extracts using a phosphosite-specific antibody [11], and indirect evidence for the phosphorylation of T248 in the human brain was obtained using a proximity ligation assay with anti-E2F4 and anti-phosphothreonine antibodies [12]. Hsu and collaborators [18] also found evidence of phosphorylation in S16, Y139, S185, S187, S220, S223, and Y389 from human E2F4 and in S75, Y139, T153, S223, S240, T266, Y392, and Y394 from mouse E2F4.
4. E2F4 as a Multifactorial Regulator
4.1. E2F4 as a Regulator of Tissue Homeostasis
In addition to its classical function in regulating quiescence in proliferating cells, E2F4 can also participate in several homeostatic processes. For instance, E2F4 has been associated with the DNA damage checkpoint and repair pathways [28][52][53] (see below), prevention of DNA damage-associated cell death [30], repression of apoptotic genes [54], epigenetics [55], metabolism regulation [56][57], autophagy [58], inflammation [59], and cell repair [60]. In addition, E2F4 function has been associated with oxidative stress [61]. In this regard, the p107-E2F4 complex downregulates PGC-1alpha expression [62], an enzyme that protects cells against oxidative stress and reduces mitochondrial dysfunction in AD [63][64].
The ability of E2F4 to regulate several homeostatic functions may have evolved from its capacity to regulate processes primarily associated with cell cycle arrest and cell differentiation. Indeed, under growth arrest conditions, E2F4 can repress a common set of genes involved in mitochondrial biogenesis and metabolism [65]. Moreover, E2F4 participates in the differentiation of multiple cell types, including the differentiation of myocytes [21][35][66][67][68], neural cells [29][69], adipocytes [70][71][72][73], hematopoietic cells [74], chondrocytes [75], extra-embryonic tissues [76], endothelial cells [77], epithelial cells [78], and multiciliated cells [79][80]. E2F4 can also regulate eye and brain patterning [81][82][83][84], as well as endocytosis and water channel transport in the testes [80].
The capacity of E2F4 to act as a multifactorial factor is likely mediated by the different interactors to which this molecule can bind. In this regard, E2F4 can perform non-canonical actions in cells in the absence of RB family proteins, allowing the transactivation domain to interact with other proteins [18]. After performing biological process (BP) ontology analysis, researchers found that many E2F4 interactors identified by these authors are related to non-cell cycle processes, including DNA repair, stem cell population maintenance, protein sumoylation in mESCs, as well as retina homeostasis, RNA splicing, organ regeneration, and regulation of lipid kinase activity in RPE-derived cells.
4.2. E2F4 as a Regulator of DNA Repair
Cells have to constantly respond to genotoxic insults that may induce DNA modifications, which usually lead to genome instability. Accumulation of damaged DNA is deleterious for cells since it often results in abnormal proliferation, cell aging, or cell death. Eukaryotic cells have acquired mechanisms of defense against this damage; globally, they are referred to as DNA damage response (DDR), which are in charge of monitoring and removing lesions in their DNA [85]. In this regard, mammalian cells are equipped with several DNA repair pathways, which can be classified into two main groups [86]. On the one hand, the machinery involved in base excision repair, nucleotide excision repair (NER), and mismatch repair can fix single-strand mutations. On the other hand, double strand breaks (DSBs) can be repared through two main mechanisms: homologous recombination (HR), which repairs DSBs during the S-phase or G2 since the sister chromatic is used as a template, and non-homologous end-joining (NHEJ), which is able to repair DSBs at any stage of the cell cycle and in quiescent and postmitotic cells.
DDR can be transcriptionally regulated by E2F factors. These transcription factors usually bind to two adjacent E2F sites within the regulatory regions of genes involved in DNA damage checkpoint and repair [87], thus allowing for functional interactions. Two known E2F factors regulating DDR are E2F4 and E2F1 [26][28], which functionally counteract each other. For instance, E2F4 silencing in MCF7 epithelial breast cells treated with benzoapyrene, an environmental pollutant that triggers DNA damage [88], results in E2F1 derepression and the subsequent induction of DNA repair factors [89]. In primary neurons, the repair response to DSBs is also regulated by E2F1 and E2F4. In this cellular system, E2F1 enhances Cited2 expression, a pro-apoptotic gene required for delayed neuronal cell death, while E2F4 strongly inhibits Cited2 transcription, helping to cell survival [30]. Finally, E2F4 has also been implicated in NER since the p130/E2F4 complex controls the expression of xeroderma pigmentosum complementation group C [52], a protein that serves as the primary initiating factor in the global genome NER pathway [90]. There is also evidence that hypoxia and the anti-angiogenic agent cediranib are both able to induce the binding of p130/E2F4 complexes to E2F consensus sequences in the promoters of homology-directed DNA repair genes, thus reducing gene expression [53][87][91].
In most paradigms, E2F4 seems to act as a repressor of genes involved in DNA damage checkpoint and repair. This function may be favored by the stress kinase p38MAPK, which phosphorylates E2F4 [13] and becomes activated by the DDR [92]. Therefore, the expression of a non-phosphorylatable form of E2F4 (E2F4DN) might modulate the maintenance of the expression of genes involved in DDR.
4.3. E2F4 as a Putative Regulator of Synaptic Function
E2F4 has been related to cognitive impairment [93] and the pathogenesis of AD [94], as well as to other neurological diseases [95], including Parkinson´s disease/mild cognitive impairment [96]. Since AD is largely a synaptic failure [97] occurring prior to cognitive decline or cell death [98], it can be speculated that E2F4 is important for synaptic function.
4.3.1. Transcriptional Regulation of Synaptic Function by E2F4
E2F4 has the potential to regulate the expression of an ample number of synaptic proteins. As evidenced by ChIP-seq datasets from the ENCODE transcription factor targets dataset interrogated with the Harmonizome tool [99], E2F4 can bind to 46 synaptic protein-encoding genes, as well as 127 genes encoding for ion channel subunits. In this regard, there is direct evidence that E2F4 can regulate synaptic function, coming from the transcriptomic analysis performed in mESCs subjected to E2f4 gene knock-out (KO). The transcriptional alterations in synaptic plasticity-related genes upon E2F4 modulation reveal the potential role of this protein in synaptic function. This suggests that E2F4 could be a promising target for several neurological diseases that course with synaptic plasticity impairment, such as AD.
4.3.2. Interaction of E2F4 with Synaptic Regulators
E2F4 can interact with synaptic regulators. Researchers verified using BP ontology that almost half of the E2F4 interactors found in the study by Hsu and collaborators [18], which are common in both mESCs and RPE-derived cells, have a function in either axonal transport or synapse physiology.
The E2F4 interactors involved in synaptic function that were identified in RPE-derived cells include Rac Family Small GTPase 1 (Rac1), cell division cycle 42 (Cdc42), and protein phosphatase 1 catalytic subunit β (PPP1CB) [18]. The actin regulators Rac1 and Cdc42 are important for the structural and functional plasticity of dendritic spines, which are the basis of learning mechanisms [100]. The actin cytoskeleton regulator Rac1 controls synaptic actin dynamics [101] and is involved in actin-regulated short-term presynaptic plasticity through the modulation of synaptic vesicle replenishment [102]. Cdc42 is known to have an important role in dendritic branching [103], and it is part of the mechanism involved in CaMKII activation, which modulates dendritic spine structural plasticity and induces LTP [104]. PPP1CB is one of the three catalytic subunits of protein phosphatase 1 (PP1), a serine/threonine protein phosphatase that regulates synaptic transmission and plasticity [105]. PP1 mediates NMDAR dephosphorylation, modulating the synaptic expression of this receptor [106].
Hsu and collaborators [18] also found Fragile X Mental Retardation Protein (FMRP) to be a candidate cofactor for E2F4 in mESCs. FMRP is an important regulator of activity-dependent plasticity in the brain, and the mutation in the FMR1 gene and subsequent loss of its protein product lead to Fragile X Syndrome (FXS), an inherited cause of autism and intellectual disability [107]. Mechanistically, FMRP is an RNA-binding protein that regulates the synthesis of synaptic and nuclear proteins within various compartments of the neuron [108]. FMRP binds to dendritic mRNA [109], and this may be important in mRNA localization to dendrites [110]. Thus, the hypothetical interaction of E2F4 with FMRP could be responsible for the modulation of synaptic protein transduction.
Hsu and collaborators [18] also found that Snapin, a protein related to synaptic function [111][112], can interact with E2F4 in both mESC and RPE cells.
In addition, the indirect effects of E2F4 on synaptic plasticity have also been described. In this regard, E2F4 can interact with Suv39H1 [113], a histone methyl transferase with an essential role in H3K9me3 methylation that mediates hippocampal memory functions [114].
The interaction of E2F4 with known synaptic regulators suggests that it may modulate synaptic function. This hypothesis is consistent with the observed enrichment of E2F1 in synaptic fractions, which is related to PSD95 expression and becomes upregulated with aging [115]. Furthermore, E2F1 is necessary for de novo neuronal tetraploidization occurring in mice, and this is associated with the alteration of cognition, as mice lacking this transcription factor show enhanced memory acquisition and consolidation [116]. Since E2F1 and E2F4 have antagonistic roles in neuronal function [95], researchers speculate that E2F4 could facilitate synaptic function and cognition, as opposed to E2F1.
4.3.3. E2F4 and MAPK Proteins in Synaptic Function
Another piece of evidence for the putative capacity of E2F4 to regulate synaptic function comes from the study by [34], which showed that E2F4 can regulate genes involved in the MAPK signaling pathway. Although this pathway has been associated with cancer [34], it is also relevant for synaptic plasticity and AD [117][118][119]. A relevant member of the MAPK family of protein kinases is p38MAPK, the kinase that phosphorylates E2F4 in the Thr conserved motif controlling neuronal tetraploidization [13]. p38MAPK is a protein involved in synaptic plasticity and memory impairment that has been widely related to AD [119][120]. Accordingly, p38MAPK is progressively activated in neurons affected by AD [121] as well as in APP transgenic mice brains [120], and neuronal p38αMAPK mediates synaptic and cognitive dysfunction in a murine model of AD by controlling amyloid-β (Aβ) production [119]. Moreover, downregulation in APP/Tau-transgenic mice of p38MAPK results in the upregulation of genes involved in the MAPK pathway and calcium signaling [120]. Although the implication of E2F4 in this paradigm remains unclear, the expression of some calcium signaling and/or synaptic plasticity-related genes is altered upon p38α-MAPK deficiency in neuronal populations. In particular, the expression of both Grin2a and its encoded protein glutamate ionotropic receptor NMDA type subunit 2A (Grin2a) is decreased, resulting in a reduction of calcium influx in p38α-MAPK-deficient neurons [120]. Finally, knocking down E2f4 using an E2f4-specific shRNA significantly decreased the protein levels of p-ERK [34], a key MAPK that has been involved in both neurodegenerative diseases, as well as in endocannabinoid [122][123][124][125][126][127] and calcium signaling [100][104][128][129][130][131][132], which are critical pathways in synaptic function and modulation.
References
- Pierce, A.M.; Schneider-Broussard, R.; Philhower, J.L.; Johnson, D.G. Differential activities of E2F family members: Unique functions in regulating transcription. Mol. Carcinog. 1998, 22, 190–198.
- Ginsberg, D.; Vairo, G.; Chittenden, T.; Xiao, Z.X.; Xu, G.; Wydner, K.L.; DeCaprio, J.A.; Lawrence, J.B.; Livingston, D.M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994, 8, 2665–2679.
- Beijersbergen, R.L.; Kerkhoven, R.M.; Zhu, L.; Carlée, L.; Voorhoeve, P.M.; Bernards, R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994, 8, 2680–2690.
- Vairo, G.; Livingston, D.M.; Ginsberg, D. Functional interaction between E2F-4 and p130: Evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995, 9, 869–881.
- Garneau, H.; Paquin, M.C.; Carrier, J.C.; Rivard, N. E2F4 expression is required for cell cycle progression of normal intestinal crypt cells and colorectal cancer cells. J. Cell Physiol. 2009, 221, 350–358.
- van Amerongen, M.J.; Diehl, F.; Novoyatleva, T.; Patra, C.; Engel, F.B. E2F4 is required for cardiomyocyte proliferation. Cardiovasc. Res. 2010, 86, 92–102.
- Crosby, M.E.; Almasan, A. Opposing roles of E2Fs in cell proliferation and death. Cancer Biol. Ther. 2004, 3, 1208–1211.
- Hsu, J.; Sage, J. Novel functions for the transcription factor E2F4 in development and disease. Cell Cycle 2016, 15, 3183–3190.
- Miles, S.; Breeden, L. A common strategy for initiating the transition from proliferation to quiescence. Curr. Genet. 2017, 63, 179–186.
- Stevaux, O.; Dyson, N.J. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 2002, 14, 684–691.
- López-Sánchez, N.; Garrido-García, A.; Ramón-Landreau, M.; Cano-Daganzo, V.; Frade, J.M. E2F4-based gene therapy mitigates the phenotype of the Alzheimer’s disease mouse model 5xFAD. Neurotherapeutics 2021, 18, 2484–2503.
- López-Sánchez, N.; Ramón-Landreau, M.; Trujillo, C.; Garrido-García, A.; Frade, J.M. A mutant variant of E2F4 triggers multifactorial therapeutic effects in 5xFAD mice. Mol. Neurobiol. 2022, 59, 3016–3039.
- Morillo, S.M.; Abanto, E.P.; Román, M.J.; Frade, J.M. Nerve growth factor-induced cell cycle reentry in newborn neurons is triggered by p38MAPK-dependent E2F4 phosphorylation. Mol. Cell. Biol. 2012, 32, 2722–2737.
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 2006, 26, 10129–10140.
- Lang, S.E.; McMahon, S.B.; Cole, M.D.; Hearing, P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 2001, 276, 32627–32634.
- Verona, R.; Moberg, K.; Estes, S.; Starz, M.; Vernon, J.P.; Lees, J.A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol. Cell. Biol. 1997, 17, 7268–7282.
- Gaubatz, S.; Lees, J.A.; Lindeman, G.J.; Livingston, D.M. E2F4 is exported from the nucleus in a CRM1-dependent manner. Mol. Cell. Biol. 2001, 21, 1384–1392.
- Hsu, J.; Arand, J.; Chaikovsky, A.; Mooney, N.A.; Demeter, J.; Brison, C.M.; Oliverio, R.; Vogel, H.; Rubin, S.M.; Jackson, P.K.; et al. E2F4 regulates transcriptional activation in mouse embryonic stem cells independently of the RB family. Nat. Commun. 2019, 10, 2939.
- Chook, Y.M.; Blobel, G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 2001, 11, 703–715.
- Avis, J.M.; Clarke, P.R. Ran, a GTPase involved in nuclear processes: Its regulators and effectors. J. Cell Sci. 1996, 109 Pt 10, 2423–2427.
- Puri, P.L.; Cimino, L.; Fulco, M.; Zimmerman, C.; La Thangue, N.B.; Giordano, A.; Graessmann, A.; Levrero, M. Regulation of E2F4 mitogenic activity during terminal differentiation by its heterodimerization partners for nuclear translocation. Cancer Res. 1998, 58, 1325–1331.
- Lindeman, G.J.; Gaubatz, S.; Livingston, D.M.; Ginsberg, D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc. Natl. Acad. Sci. USA 1997, 94, 5095–5100.
- Magae, J.; Wu, C.L.; Illenye, S.; Harlow, E.; Heintz, N.H. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J. Cell Sci. 1996, 109, 1717–1726.
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shutling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176.
- Apostolova, M.D.; Ivanova, I.A.; Dagnino, C.; D’Souza, S.J.; Dagnino, L. Active nuclear import and export pathways regulate E2F-5 subcellular localization. J. Biol. Chem. 2002, 277, 34471–34479.
- Lee, B.K.; Bhinge, A.A.; Iyer, V.R. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucl. Acids Res. 2011, 39, 3558–3573.
- Xu, X.; Bieda, M.; Jin, V.X.; Rabinovich, A.; Oberley, M.J.; Green, R.; Farnham, P.J. A comprehensive ChIP-chip analysis of E2F1; E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007, 17, 1550–1561.
- Ren, B.; Cam, H.; Takahashi, Y.; Volkert, T.; Terragni, J.; Young, R.A.; Dynlacht, B.D. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002, 16, 245–256.
- Julian, L.M.; Liu, Y.; Pakenham, C.A.; Dugal-Tessier, D.; Ruzhynsky, V.; Bae, S.; Tsai, S.Y.; Leone, G.; Slack, R.S.; Blais, A. Tissue-specific targeting of cell fate regulatory genes by E2f factors. Cell Death Differ. 2016, 23, 565–575.
- Huang, T.; González, Y.R.; Qu, D.; Huang, E.; Safarpour, F.; Wang, E.; Joselin, A.; Im, D.S.; Callaghan, S.M.; Boonying, W.; et al. The pro-death role of Cited2 in stroke is regulated by E2F1/4 transcription factors. J. Biol. Chem. 2019, 294, 8617–8629.
- Rabinovich, A.; Jin, V.X.; Rabinovich, R.; Xu, X.; Farnham, P.J. E2F in vivo binding specificity: Comparison of consensus versus nonconsensus binding sites. Genome Res. 2008, 18, 1763–1777.
- Tommasi, S.; Pfeifer, G.P. In vivo structure of the human cdc2 promoter: Release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 1995, 15, 6901–6913.
- Tuncay, K.; Ensman, L.; Sun, J.; Haidar, A.A.; Stanley, F.; Trelinski, M.; Ortoleva, P. Transcriptional regulatory networks via gene ontology and expression data. In Silico Biol. 2007, 7, 21–34.
- Feng, Y.; Li, L.; Du, Y.; Peng, X.; Chen, F. E2F4 functions as a tumour suppressor in acute myeloid leukaemia via inhibition of the MAPK signalling pathway by binding to EZH2. J. Cell. Mol. Med. 2020, 24, 2157.
- Gill, R.M.; Hamel, P.A. Subcellular compartmentalization of E2F family members is required for maintenance of the postmitotic state in terminally differentiated muscle. J. Cell Biol. 2000, 148, 1187–1201.
- Hazan, R.; Mori, M.; Danielian, P.S.; Guen, V.J.; Rubin, S.M.; Cardoso, W.V.; Lees, J.A. E2F4’s cytoplasmic role in multiciliogenesis is mediated via an N-terminal domain that binds two components of the centriole replication machinery, Deup1 and SAS6. Mol. Biol. Cell 2021, 32, ar1.
- Mori, M.; Hazan, R.; Danielian, P.S.; Mahoney, J.E.; Li, H.; Lu, J.; Miller, E.S.; Zhu, X.; Lees, J.A.; Cardoso, W.V. Cytoplasmic E2f4 forms organizing centres for initiation of centriole amplification during multiciliogenesis. Nat. Commun. 2017, 8, 15857.
- Stracker, T.H. E2F4/5-mediated transcriptional control of multiciliated cell differentiation: Redundancy or fine-tuning? Dev. Biol. 2019, 446, 20–21.
- Ma, L.; Quigley, I.; Omran, H.; Kintner, C. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 2014, 28, 1461–1471.
- Chong, Y.L.; Zhang, Y.; Zhou, F.; Roy, S. Distinct requirements of E2f4 versus E2f5 activity for multiciliated cell development in the zebrafish embryo. Dev. Biol. 2018, 443, 165–172.
- Ramazi, S.; Zahiri, J. Post-translational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012.
- Khoury, G.A.; Baliban, R.C.; Floudas, C.A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011, 1, 90.
- Wang, S.A.; Wang, Y.C.; Chuang, Y.P.; Huang, Y.H.; Su, W.C.; Chang, W.C.; Hung, J.J. EGF-mediated inhibition of ubiquitin-specific peptidase 24 expression has a crucial role in tumorigenesis. Oncogene 2017, 36, 2930–2945.
- Martínez-Balbás, M.A.; Bauer, U.M.; Nielsen, S.J.; Brehm, A.; Kouzarides, T. Regulation of E2F1 activity by acetylation. EMBO J. 2000, 19, 662–671.
- Carr, S.M.; Poppy Roworth, A.; Chan, C.; La Thangue, N.B. Post-translational control of transcription factors: Methylation ranks highly. FEBS J. 2015, 282, 4450–4465.
- Kontaki, H.; Talianidis, I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell 2010, 39, 152–160.
- Xie, Q.; Bai, Y.; Wu, J.; Sun, Y.; Wang, Y.; Zhang, Y.; Mei, P.; Yuan, Z. Methylation-mediated regulation of E2F1 in DNA damage-induced cell death. J. Recept. Signal Transduct. Res. 2011, 31, 139–146.
- Advani, S.J.; Weichselbaum, R.R.; Roizman, B. E2F proteins are posttranslationally modified concomitantly with a reduction in nuclear binding activity in cells infected with herpes simplex virus 1. J Virol. 2000, 74, 7842–7850.
- Scimè, A.; Li, L.; Ciavarra, G.; Whyte, P. Cyclin D1/cdk4 can interact with E2F4/DP1 and disrupts its DNA-binding capacity. J. Cell Physiol. 2008, 214, 568–581.
- Araki, K.; Kawauchi, K.; Tanaka, N. IKK/NF-kappaB signaling pathway inhibits cell-cycle progression by a novel Rb-independent suppression system for E2F transcription factors. Oncogene 2008, 27, 5696–5705.
- Paquin, M.C.; Cagnol, S.; Carrier, J.C.; Leblanc, C.; Rivard, N. ERK-associated changes in E2F4 phosphorylation, localization and transcriptional activity during mitogenic stimulation in human intestinal epithelial crypt cells. BMC Cell Biol. 2013, 14, 33.
- Dominguez-Brauer, C.; Chen, Y.J.; Brauer, P.M.; Pimkina, J.; Raychaudhuri, P. ARF stimulates XPC to trigger nucleotide excision repair by regulating the repressor complex of E2F4. EMBO Rep. 2009, 10, 1036–1042.
- Kaplan, A.R.; Gueble, S.E.; Liu, Y.; Oeck, S.; Kim, H.; Yun, Z.; Glazer, P.M. Cediranib suppresses homology-directed DNA repair through down-regulation of BRCA1/2 and RAD51. Sci. Transl. Med. 2019, 11, eaav4508.
- Dingar, D.; Konecny, F.; Zou, J.; Sun, X.; von Harsdorf, R. Anti-apoptotic function of the E2F transcription factor 4 (E2F4)/p130, a member of retinoblastoma gene family in cardiac myocytes. J. Mol. Cell. Cardiol. 2012, 53, 820–828.
- Luo, C.; Sheng, J.; Hu, M.G.; Haluska, F.G.; Cui, R.; Xu, Z.; Tsichlis, P.N.; Hu, G.F.; Hinds, P.W. Loss of ARF sensitizes transgenic BRAFV600E mice to UV-induced melanoma via suppression of XPC. Cancer Res. 2013, 73, 4337–4348.
- Pamuklar, Z.N.; Chen, J.; Muehlbauer, M.; Spagnoli, A.; Torquati, A. Necdin-E2F4 interaction provides insulin-sensitizing effect after weight loss induced by gastric bypass surgery. Surg. Obes. Relat. Dis. 2013, 9, 94–99.
- Zhao, Z.D.; Zan, L.S.; Li, A.N.; Cheng, G.; Li, S.J.; Zhang, Y.R.; Wang, X.Y.; Zhang, Y.Y. Characterization of the promoter region of the bovine long-chain acyl-CoA synthetase 1 gene: Roles of E2F1, Sp1, KLF15, and E2F4. Sci. Rep. 2016, 6, 19661.
- Sutton, M.N.; Huang, G.Y.; Zhou, J.; Mao, W.; Langley, R.; Lu, Z.; Bast, R.C., Jr. Amino Acid Deprivation-Induced Autophagy Requires Upregulation of DIRAS3 through Reduction of E2F1 and E2F4 Transcriptional Repression. Cancers 2019, 11, 603.
- Petrenko, O.; Moll, U.M. Macrophage migration inhibitory factor MIF interferes with the Rb-E2F pathway. Mol. Cell 2005, 17, 225–236.
- Zhang, X.; Chen, J.; Xue, M.; Tang, Y.; Xu, J.; Liu, L.; Huang, Y.; Yang, Y.; Qiu, H.; Guo, F. Overexpressing p130/E2F4 in mesenchymal stem cells facilitates the repair of injured alveolar epithelial cells in LPS-induced ARDS mice. Stem Cell Res. Ther. 2019, 10, 74.
- Zhu, B.; Khozoie, C.; Bility, M.T.; Ferry, C.H.; Blazanin, N.; Glick, A.B.; Gonzalez, F.J.; Peters, J.M. Peroxisome proliferator-activated receptor β/δ cross talks with E2F and attenuates mitosis in HRAS-expressing cells. Mol. Cell. Biol. 2012, 32, 2065–2082.
- Scimè, A.; Soleimani, V.D.; Bentzinger, C.F.; Gillespie, M.A.; Le Grand, F.; Grenier, G.; Bevilacqua, L.; Harper, M.E.; Rudnicki, M.A. Oxidative status of muscle is determined by p107 regulation of PGC-1alpha. J. Cell Biol. 2010, 190, 651–662.
- Qin, W.; Haroutunian, V.; Katsel, P.; Cardozo, C.P.; Ho, L.; Buxbaum, J.D.; Pasinetti, G.M. PGC-1alpha expression decreases in the Alzheimer disease brain as a function of dementia. Arch. Neurol. 2009, 66, 352–361.
- Sweeney, G.; Song, J. The association between PGC-1α and Alzheimer’s disease. Anat. Cell Biol. 2016, 49, 1–6.
- Cam, H.; Balciunaite, E.; Blais, A.; Spektor, A.; Scarpulla, R.C.; Young, R.; Kluger, Y.; Dynlacht, B.D. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 2004, 16, 399–411.
- Puri, P.L.; Balsano, C.; Burgio, V.L.; Chirillo, P.; Natoli, G.; Ricci, L.; Mattei, E.; Graessmann, A.; Levrero, M. MyoD prevents cyclinA/cdk2 containing E2F complexes formation in terminally differentiated myocytes. Oncogene 1997, 14, 1171–1184.
- Parakati, R.; DiMario, J.X. Dynamic transcriptional regulatory complexes, including E2F4, p107, p130, and Sp1, control fibroblast growth factor receptor 1 gene expression during myogenesis. J. Biol. Chem. 2005, 280, 21284–21294.
- An, Y.; Wang, G.; Diao, Y.; Long, Y.; Fu, X.; Weng, M.; Zhou, L.; Sun, K.; Cheung, T.H.; Ip, N.Y.; et al. A molecular switch regulating cell fate choice between muscle progenitor cells and brown adipocytes. Dev. Cell 2017, 41, e5–e391.
- Persengiev, S.P.; Kondova, I.I.; Kilpatrick, D.L. E2F4 actively promotes the initiation and maintenance of nerve growth factor-induced cell differentiation. Mol. Cell Biol. 1999, 19, 6048–6056.
- Fajas, L.; Landsberg, R.L.; Huss-Garcia, Y.; Sardet, C.; Lees, J.A.; Auwerx, J. E2Fs regulate adipocyte differentiation. Dev. Cell 2002, 3, 39–49.
- Landsberg, R.L.; Sero, J.E.; Danielian, P.S.; Yuan, T.L.; Lee, E.Y.; Lees, J.A. The role of E2F4 in adipogenesis is independent of its cell cycle regulatory activity. Proc. Natl. Acad. Sci. USA 2003, 100, 2456–2461.
- Miard, S.; Fajas, L. Atypical transcriptional regulators and cofactors of PPARgamma. Int. J. Obes. 2005, 29 (Suppl. S1), S10–S12.
- Tseng, Y.H.; Butte, A.J.; Kokkotou, E.; Yechoor, V.K.; Taniguchi, C.M.; Kriauciunas, K.M.; Cypess, A.M.; Niinobe, M.; Yoshikawa, K.; Patti, M.E.; et al. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat. Cell Biol. 2005, 7, 601–611.
- Enos, M.E.; Bancos, S.A.; Bushnell, T.; Crispe, I.N. E2F4 modulates differentiation and gene expression in hematopoietic progenitor cells during commitment to the lymphoid lineage. J. Immunol. 2008, 180, 3699–3707.
- Yanagino, T.; Yuasa, K.; Nagahama, M.; Matsuda, Y.; Tsuji, A. Transcriptional regulation of fibrillin-2 gene by E2F family members in chondrocyte differentiation. J. Cell. Biochem. 2009, 106, 580–588.
- Lee, E.Y.; Yuan, T.L.; Danielian, P.S.; West, J.C.; Lees, J.A. E2F4 cooperates with pRB in the development of extra-embryonic tissues. Dev. Biol. 2009, 332, 104–115.
- Gu, L.; Hitzel, J.; Moll, F.; Kruse, C.; Malik, R.A.; Preussner, J.; Looso, M.; Leisegang, M.S.; Steinhilber, D.; Brandes, R.P.; et al. The histone demethylase PHF8 is essential for endothelial cell migration. PLoS ONE 2016, 11, e0146645.
- Zhen, Z.; Zhang, M.; Yuan, X.; Li, M. Transcription factor E2F4 is a positive regulator of milk biosynthesis and proliferation of bovine mammary epithelial cells. Cell Biol. Int. 2020, 44, 229–241.
- Danielian, P.S.; Bender Kim, C.F.; Caron, A.M.; Vasile, E.; Bronson, R.T.; Lees, J.A. E2f4 is required for normal development of the airway epithelium. Dev. Biol. 2007, 305, 564–576.
- Danielian, P.S.; Hess, R.A.; Lees, J.A. E2f4 and E2f5 are essential for the development of the male reproductive system. Cell Cycle 2016, 15, 250–260.
- Kastner, A.; Espanel, X.; Brun, G. Transient accumulation of retinoblastoma/E2F-1 protein complexes correlates with the onset of neuronal differentiation in the developing quail neural retina. Cell Growth Differ. 1998, 9, 857–867.
- Francesconi, C.M.; Hutcheon, A.E.; Chung, E.H.; Dalbone, A.C.; Joyce, N.C.; Zieske, J.D. Expression patterns of retinoblastoma and E2F family proteins during corneal development. Investig. Ophthalmol. Vis. Sci. 2000, 41, 1054–1062.
- Ruzhynsky, V.A.; McClellan, K.A.; Vanderluit, J.L.; Jeong, Y.; Furimsky, M.; Park, D.S.; Epstein, D.J.; Wallace, V.A.; Slack, R.S. Cell cycle regulator E2F4 is essential for the development of the ventral telencephalon. J. Neurosci. 2007, 27, 5926–5935.
- Ruzhynsky, V.A.; Furimsky, M.; Park, D.S.; Wallace, V.A.; Slack, R.S. E2F4 is required for early eye patterning. Dev. Neurosci. 2009, 31, 238–246.
- Yao, S.; Feng, Y.; Zhang, Y.; Feng, J. DNA damage checkpoint and repair: From the budding yeast Saccharomyces cerevisiae to the pathogenic fungus Candida albicans. Comput. Struct. Biotechnol. J. 2021, 19, 6343–6354.
- Huang, R.; Zhou, P.K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target Ther. 2021, 6, 254.
- Bindra, R.S.; Gibson, S.L.; Meng, A.; Westermark, U.; Jasin, M.; Pierce, A.J.; Bristow, R.G.; Classon, M.K.; Glazer, P.M. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005, 65, 11597–11604.
- Bukowska, B.; Mokra, K.; Michałowicz, J. Benzopyrene-Environmental Occurrence, Human Exposure, and Mechanisms of Toxicity. Int. J. Mol. Sci. 2022, 23, 6348.
- Allmann, S.; Mayer, L.; Olma, J.; Kaina, B.; Hofmann, T.G.; Tomicic, M.T.; Christmann, M. Benzopyrene represses DNA repair through altered E2F1/E2F4 function marking an early event in DNA damage-induced cellular senescence. Nucl. Acids Res. 2020, 48, 12085–12101.
- Shell, S.M.; Hawkins, E.K.; Tsai, M.S.; Hlaing, A.S.; Rizzo, C.J.; Chazin, W.J. Xeroderma pigmentosum complementation group C protein (XPC) serves as a general sensor of damaged DNA. DNA Repair 2013, 12, 947–953.
- Bindra, R.S.; Glazer, P.M. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene 2007, 26, 2048–2057.
- Lee, S.; Lee, H.S.; Baek, M.; Lee, D.Y.; Bang, Y.J.; Cho, H.N.; Lee, Y.S.; Ha, J.H.; Kim, H.Y.; Jeoung, D.I. MAPK signaling is involved in camptothecin-induced cell death. Mol. Cells 2002, 14, 348–354.
- Muiño, E.; Maisterra, O.; Jiménez-Balado, J.; Cullell, N.; Carrera, C.; Torres-Aguila, N.P.; Cárcel-Márquez, J.; Gallego-Fabrega, C.; Lledós, M.; González-Sánchez, J.; et al. Genome-wide transcriptome study in skin biopsies reveals an association of E2F4 with cadasil and cognitive impairment. Sci. Rep. 2021, 11, 6846.
- Ding, J.; Kong, W.; Mou, X.; Wang, S. Construction of Transcriptional Regulatory Network of Alzheimer’s Disease Based on PANDA Algorithm. Interdiscip. Sci. 2019, 11, 226–236.
- Iyirhiaro, G.O.; Zhang, Y.; Estey, C.; O’Hare, M.J.; Safarpour, F.; Parsanejad, M.; Wang, S.; Abdel-Messih, E.; Callaghan, S.M.; During, M.J.; et al. Regulation of ischemic neuronal death by E2F4-p130 protein complexes. J. Biol. Chem. 2014, 289, 18202–18213.
- Jiang, Z.; Huang, Y.; Zhang, P.; Han, C.; Lu, Y.; Mo, Z.; Zhang, Z.; Li, X.; Zhao, S.; Cai, F.; et al. Characterization of a pathogenic variant in GBA for Parkinson’s disease with mild cognitive impairment patients. Mol. Brain 2020, 13, 102.
- Selkoe, D.J. Alzheimer’s disease is a synaptic failure. Science 2002, 298, 789–791.
- Knafo, S.; Sánchez-Puelles, C.; Palomer, E.; Delgado, I.; Draffin, J.E.; Mingo, J.; Wahle, T.; Kaleka, K.; Mou, L.; Pereda-Perez, I.; et al. PTEN recruitment controls synaptic and cognitive function in Alzheimer’s models. Nat. Neurosci. 2016, 19, 443–453.
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100.
- Tu, X.; Yasuda, R.; Colgan, L.A. Rac1 is a downstream effector of PKCα in structural synaptic plasticity. Sci. Rep. 2020, 10, 1777.
- Lo, L.H.Y.; Dong, R.; Lyu, Q.; Lai, K.O. The Protein Arginine Methyltransferase PRMT8 and Substrate G3BP1 Control Rac1-PAK1 Signaling and Actin Cytoskeleton for Dendritic Spine Maturation. Cell Rep. 2020, 31, 107744.
- O’Neil, S.D.; Rácz, B.; Brown, W.E.; Gao, Y.; Soderblom, E.J.; Yasuda, R.; Soderling, S.H. Action potential-coupled Rho GTPase signaling drives presynaptic plasticity. eLife 2021, 10, e63756.
- Gauthier-Campbell, C.; Bredt, D.S.; Murphy, T.H.; El-Din El-Husseini, A. Regulation of Dendritic Branching and Filopodia Formation in Hippocampal Neurons by Specific Acylated Protein Motifs. Mol. Biol. Cell 2004, 15, 2205–2217.
- Shibata, A.; Ueda, H.H.; Eto, K.; Onda, M.; Sato, A.; Ohba, T.; Nabekura, J.; Murakoshi, H. Photoactivatable CaMKII induces synaptic plasticity in single synapses. Nat. Commun. 2021, 12, 751.
- Foley, K.; McKee, C.; Nairn, A.C.; Xia, H. Regulation of Synaptic Transmission and Plasticity by Protein Phosphatase 1. J. Neurosci. 2021, 41, 3040–3050.
- Chiu, A.M.; Wang, J.; Fiske, M.P.; Hubalkova, P.; Barse, L.; Gray, J.A.; Sanz-Clemente, A. NMDAR-Activated PP1 Dephosphorylates GluN2B to Modulate NMDAR Synaptic Content. Cell Rep. 2019, 28, e5–e341.
- Liu, X.; Kumar, V.; Tsai, N.P.; Auerbach, B.D. Hyperexcitability and Homeostasis in Fragile X Syndrome. Front. Mol. Neurosci. 2022, 14, 805929.
- Yoon, Y.J.; Bassell, G.J. Diversity on location. eLife 2022, 11, e76818.
- Hale, C.R.; Sawicka, K.; Mora, K.; Fak, J.J.; Kang, J.J.; Cutrim, P.; Cialowicz, K.; Carroll, T.S.; Darnell, R.B. FMRP regulates mRNAs encoding distinct functions in the cell body and dendrites of CA1 pyramidal neurons. eLife 2021, 10, e71892.
- Zalfa, F.; Eleuteri, B.; Dickson, K.S.; Mercaldo, V.; De Rubeis, S.; di Penta, A.; Tabolacci, E.; Chiurazzi, P.; Neri, G.; Grant, S.G.; et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat. Neurosci. 2007, 10, 578–587.
- Schnöder, L.; Tomic, I.; Schwindt, L.; Helm, D.; Rettel, M.; Schulz-Schaeffer, W.; Krause, E.; Rettig, J.; Fassbender, K.; Liu, Y. P38α-MAPK phosphorylates Snapin and reduces Snapin-mediated BACE1 transportation in APP-transgenic mice. FASEB J. 2021, 35, e21691.
- Ye, X.; Feng, T.; Tammineni, P.; Chang, Q.; Jeong, Y.Y.; Margolis, D.J.; Cai, H.; Kusnecov, A.; Cai, Q. Regulation of Synaptic Amyloid-β Generation through BACE1 Retrograde Transport in a Mouse Model of Alzheimer’s Disease. J. Neurosci. 2017, 37, 2639–2655.
- Greene, L.A.; Liu, D.X.; Troy, C.M.; Biswas, S.C. Cell cycle molecules define a pathway required for neuron death in development and disease. Biochim. Biophys. Acta 2007, 1772, 392–401.
- Snigdha, S.; Prieto, G.A.; Petrosyan, A.; Loertscher, B.M.; Dieskau, A.P.; Overman, L.E.; Cotman, C.W. H3K9me3 inhibition improves memory, promotes spine formation, and increases BDNF levels in the aged hippocampus. J. Neurosci. 2016, 36, 3611–3622.
- Ting, J.H.; Marks, D.R.; Schleidt, S.S.; Wu, J.N.; Zyskind, J.W.; Lindl, K.A.; Blendy, J.A.; Pierce, R.C.; Jordan-Sciutto, K.L. Targeted gene mutation of E2F1 evokes age-dependent synaptic disruption and behavioral deficits. J. Neurochem. 2014, 129, 850.
- López-Sánchez, N.; Frade, J.M. : A mutant form of E2F4 prevents neuronal tetraploidization and cognitive deficits in 5xFAD mice without affecting Aβ deposition. Alzheimer’s Dement. 2017, 13, P659–P661.
- Hoerndli, F.J.; Brockie, P.J.; Wang, R.; Mellem, J.E.; Kallarackal, A.; Doser, R.L.; Pierce, D.M.; Madsen, D.M.; Maricq, A.V. MAPK signaling and a mobile scaffold complex regulate AMPA receptor transport to modulate synaptic strength. Cell Rep. 2022, 38, 110577.
- Navarrete, M.; Cuartero, M.I.; Palenzuela, R.; Draffin, J.E.; Konomi, A.; Serra, I.; Colié, S.; Castaño-Castaño, S.; Hasan, M.T.; Nebreda, Á.R.; et al. Astrocytic p38α MAPK drives NMDA receptor-dependent long-term depression and modulates long-term memory. Nat. Commun. 2019, 10, 2968.
- Colié, S.; Sarroca, S.; Palenzuela, R.; Garcia, I.; Matheu, A.; Corpas, R.; Dotti, C.G.; Esteban, J.A.; Sanfeliu, C.; Nebreda, A.R. Neuronal p38α mediates synaptic and cognitive dysfunction in an Alzheimer’s mouse model by controlling β-amyloid production. Sci. Rep. 2017, 7, 45306.
- Schnöder, L.; Gasparoni, G.; Nordström, K.; Schottek, A.; Tomic, I.; Christmann, A.; Schäfer, K.H.; Menger, M.D.; Walter, J.; Fassbender, K.; et al. Neuronal deficiency of p38α-MAPK ameliorates symptoms and pathology of APP or Tau-transgenic Alzheimer’s mouse models. FASEB J. 2020, 34, 9628–9649.
- Pei, J.J.; Braak, E.; Braak, H.; Grundke-Iqbal, I.; Iqbal, K.; Winblad, B.; Cowburn, R.F. Localization of active forms of C-jun kinase (JNK) and p38 kinase in Alzheimer’s disease brains at different stages of neurofibrillary degeneration. J. Alzheimer’s Dis. 2001, 3, 41–48.
- Misner, D.L.; Sullivan, J.M. Mechanism of cannabinoid effects on long-term potentiation and depression in hippocampal CA1 neurons. J. Neurosci. 1999, 19, 6795–6805.
- Navarrete, M.; Araque, A. Endocannabinoids potentiate synaptic transmission through stimulation of astrocytes. Neuron 2010, 68, 113–126.
- Carlson, G.; Wang, Y.; Alger, B.E. Endocannabinoids facilitate the induction of LTP in the hippocampus. Nat. Neurosci. 2002, 5, 723–724.
- Madroñal, N.; Gruart, A.; Valverde, O.; Espadas, I.; Moratalla, R.; Delgado-García, J.M. Involvement of cannabinoid CB1 receptor in associative learning and in hippocampal CA3-CA1 synaptic plasticity. Cereb. Cortex 2012, 22, 550–566.
- Robin, L.M.; Oliveira da Cruz, J.F.; Langlais, V.C.; Martin-Fernandez, M.; Metna-Laurent, M.; Busquets-Garcia, A.; Bellocchio, L.; Soria-Gomez, E.; Papouin, T.; Varilh, M.; et al. Astroglial CB1 Receptors Determine Synaptic D-Serine Availability to Enable Recognition Memory. Neuron 2018, 98, e5–e944.
- Auclair, N.; Otani, S.; Soubrie, P.; Crepelm, F. Cannabinoids modulate synaptic strength and plasticity at glutamatergic synapses of rat prefrontal cortex pyramidal neurons. J. Neurophysiol. 2000, 83, 3287–3293.
- Regehr, W.G.; Tank, D.W. Postsynaptic NMDA receptor-mediated calcium accumulation in hippocampal CA1 pyramidal cell dendrites. Nature 1990, 345, 807–810.
- Hayashi, Y.; Shi, S.H.; Esteban, J.A.; Piccini, A.; Poncer, J.C.; Malinow, R. Driving AMPA receptors into synapses by LTP and CaMKII: Requirement for GluR1 and PDZ domain interaction. Science 2000, 287, 2262–2267.
- Silva, A.J.; Stevens, C.; Tonegawa, S.; Wang, Y. Deficient hippocampal long-term potentiation in α-calcium-calmodulin kinase II mutant mice. Science 1992, 257, 201–206.
- Chang, J.Y.; Parra-Bueno, P.; Laviv, T.; Szatmari, E.M.; Lee, S.J.R.; Yasuda, R. CaMKII Autophosphorylation Is Necessary for Optimal Integration of Ca2+ Signals during LTP Induction, but Not Maintenance. Neuron 2017, 94, 800–808.
- Murakoshi, H.; Wang, H.; Yasuda, R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature 2011, 472, 100–104.
More
Information
Subjects:
Neurosciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
647
Revisions:
2 times
(View History)
Update Date:
24 Oct 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




