
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xiaoliang Zhu | + 1892 word(s) | 1892 | 2020-11-10 09:35:02 | | | |
| 2 | Vicky Zhou | Meta information modification | 1892 | 2020-11-16 09:12:32 | | |
Video Upload Options
CD4 T helper cells, including Th1, Th2, Th17, Treg and Tfh, play a central role in orchestrating adaptive immune responses.
1. Introduction
To defend countless pathogen invasions, mammals have developed and evolved a rigorous and precise immune system, which is built upon various kinds of cells, cytokines, chemokines, antibodies et al. [1][2]. CD4 T helper cells, as a key component of this system, play a critical role in defending infections throughout life by coordinating the actions of other immune cells as well as non-immune cells. CD4 T cells originate from common lymphoid progenitors in the bone marrow, develop in the thymus, and execute their functions mainly in peripheral tissues and various lymphoid organs [3]. The developmental process is precisely regulated so that CD4 T cells may efficiently recognize a very broad range of foreign antigens without eliciting auto-reactivity to self-antigens. For executing their effector functions, naïve CD4 T cells first need to be activated when receiving pathogen signals delivered by antigen-presenting cells (APCs), such as dendritic cells (DCs), and then differentiate into distinct T effector (Teff) populations based on the nature of infections. Effector CD4 T cells provide host defensive mechanisms against pathogens through the production of effector cytokines, which activate other immune cells such as macrophages and CD8 T cells in killing infected cells and help B cells in producing antibodies to mediate humoral immune responses [4][5][6].
Based on the expression of signature cytokines and lineage-specific master transcription factors, CD4 T helper (Th) cells are typically categorized into five major subsets (Figure 1): Th1 (interferon (IFN)-γ and T-bet), Th2 (interleukin (IL)-4/IL-5/IL-13 and GATA3), Th17 (IL-17/IL-22 and RORγt), Tfh (IL-21 and Bcl6) and Treg (T regulatory) (IL-10/transforming growth factor (TGF)-β/IL-35 and Foxp3) [7]. Some labs have also described some other subsets, such as Th3 (TGF-β) [8], Tr1 (IL-10) [9], Th9 (IL-9) [10][11], and Th22 (IL-22) [12].
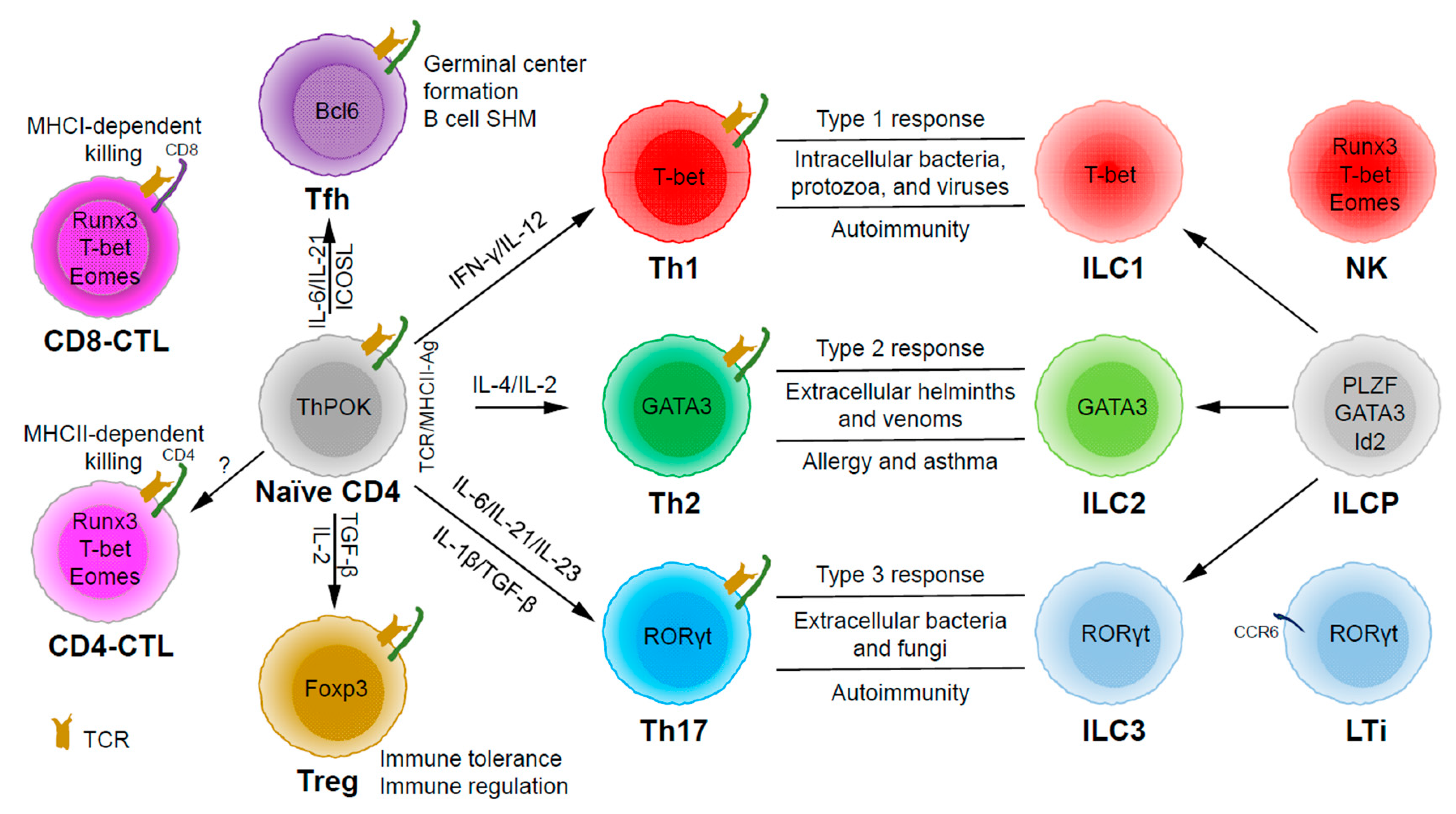
2. Functions of CD4 T Helper
Through thymic selection, CD4/CD8 double positive T cells develop into either mature CD4 (helper) or CD8 (cytotoxic) T cells, which recognize major histocompatibility complex (MHC) class II (MHCII)/peptide and MHCI/peptide complexes, respectively (Figure 1). However, some mature CD4 T cells, cytotoxic CD4 T cells (CD4-CTLs or Th-CTLs), may act similarly as cytotoxic CD8 T cells in killing their target cells through the secretion of granzyme B and perforin but in an MHCII-restricted manner [13][14][15]. The transcriptional factor Eomes (a T-box transcription factor critical for the development of functional natural killer (NK) and CD8 T cells), T-bet, and Blimp1 are critical for inducing the expression of cytotoxicity-associated genes in CD4-CTLs [16][17][18]. Similar to CD8 T cells, CD4-CTLs display increased Runx3 but reduced ThPOK expression [19][20][21][22]. Furthermore, opposite to Tfh cells, CD4-CTLs express low levels of Bcl6 but high levels of Blimp-1 [23]. Two important markers for CD4-CTLs are CRTAM [24] and NKG2C/E [25]. It has been shown that the cytotoxicity executed through CD4-CTLs becomes effective when CD8-CTL activity is impaired during infections in association with virus escape strategies [14][15]. Interestingly, more CD4-CTLs have been identified in severe COVID-19 patients than in mild symptomatic patients [26], suggesting a possible inefficient CD8 response in viral control in patients with severe disease, which is an important question that needs to be tested in the future. In addition to protecting the host from viral infections, CD4-CTLs are also involved in anti-tumor responses as well as in various autoimmune conditions [19][27][28]. Strikingly, CD4-CTLs are increased in people of exceptional longevity [29]. The development and functions of CD4-CTLs have been recently reviewed [15]. In consideration of the scope of this review, we will not further discuss this interesting population below in great detail.
Among the CD4 T subsets possessing helper functions (Figure 1), Th1 cells induce type 1 responses to protect against intracellular pathogens such as bacteria, viruses, and protozoa by activating type 1 macrophages (M1) [5]. Th2 cells are involved in type 2 responses to fight infections by parasites such as helminths by activating type 2 microphages (M2), as well as the recruitment of eosinophils, basophils, and mast cells to the sites of infections [30]. Th17 cells mediate type 3 responses for the clearance of extracellular bacteria and fungi by orchestrating sustained neutrophil recruitment and an induction of antimicrobial peptide production by epithelial cells of barrier tissues such as intestines, lungs, and skin [31]. Tfh cells are found to be important in helping B cells make antibody responses by promoting germinal center formation, affinity maturation, and possibly immunoglobulin class switch recombination [32], although Tfh-independent antibody responses have also been reported [33]. Treg cells are critical for the preservation of immune tolerance and involved in preventing autoimmune diseases [34][35].
Innate lymphoid cells (ILCs) [7][36][37] that do not express antigen receptors have been recently identified as the counterparts of T helper subsets within the innate arm of the immune system. While progenitors of ILCs are found both in bone marrow and tissue, mature ILCs are largely tissue-resident [38]. Therefore, ILCs can act earlier and more quickly than CD4 T cells during an immune response and thus provide the first line of protection against pathogens through a rapid and local secretion of effector cytokines, which were previously thought to be largely produced by CD4 T helper cells. Similar to the classification of CD4 T effector subsets into three major subsets, Th1/Th2/Th17 cells, ILCs can also be categorized into three major groups (Figure 1), which produce similar effector cytokines and lineage-defining transcription factors. Thus, group 1 ILCs (ILC1s) secret IFN-γ and express T-bet; group 2 ILCs (ILC2s) secret IL-5 and IL-13, and express GATA3; group 3 ILCs (ILC3s) secret IL-17 and IL-22, and express RORγt [7]. Functionally, group 1 ILCs, similar to Th1 cells, may participate in defending intracellular pathogens, such as bacteria, viruses, and protozoa; group 2 ILCs, just as Th2 cells, respond to large extracellular pathogens, such as parasites and allergens; group 3 ILCs, like Th17 cells, combat extracellular microbes, such as bacteria and fungi [37]. ILCs, similar to Th cells, are also involved in many inflammatory diseases including autoimmunity, allergy, and asthma [39]. It is worth pointing out that within the RORγt-expressing ILCs, there is a lymphoid tissue inducer (LTi) population, and they are involved in the formation of lymphoid structures as well as in host defense [40]. Recently, ILCs were also found to be involved in non-immunological functions including tissue repair, metabolism regulation, and immune-neuronal interactions [36].
A proper and effective response to a particular microorganism invasion through the activation and differentiation of T helper subsets and ILCs protects host from that pathogen without causing excessive inflammation that damages host tissues. However, when an immune response is out of balance, either too weak or too strong, it can cause chronic infection or inflammatory diseases [5][6][32][41][42]. Inappropriate immune responses to harmless environmental agents or self-antigens may also lead to inflammation, allergy, and autoimmunity, such as inflammatory bowel disease (IBD). Immunodeficiency with reduced ILC and CD4 T cell numbers and/or diminished ILC and CD4 T cell functions can result in insufficient immune response, which may cause chronic or repeated infections. In fact, in human immunodeficiency virus (HIV)-infected patients, the CD4 T cell number is drastically reduced, which is the main cause of acquired immunodeficiency syndrome (AIDS). X-linked severe combined immune deficiency (SCID) patients have mutations in the IL2RG gene [43], which results in a lack of T cells and ILCs; these boys suffer from severe infections of bacteria, viruses, and fungi, and they do not survive beyond infancy without treatment. In this review, we will discuss the differentiation and functions of major T helper subsets and their involvement in host defense and diseases in the context of ILCs.
In order to protect a host from the invasion of various pathogens, the immune system has evolved a series of mechanisms to respond rapidly and effectively. CD4 T helper subsets mediate effective adaptive immune responses particularly when innate immune responses fail to eliminate pathogens; CD4 T cell responses also generate memory, which can quickly respond to the second infection of the same or similar pathogens. Different types of adaptive immune responses are executed by Th1/Th2/Th17 cells, respectively, with each subset of T helper cells possessing their unique properties in producing their signature cytokines. Interestingly, before adaptive responses take place, innate lymphoid cells, which can also be divided into ILC1/ILC2/ILC3 subsets, may provide early protection and allow time for adaptive responses, whereas memory and/or memory-phenotype CD4 T cells, by possessing some similar features of ILCs (i.e., a completed differentiation process allowing a rapid response, capable of responding to inflammatory cytokines and tissue residency), may behave similarly as ILCs, to a certain extent, during a subsequent infection [44][45].
Th1, Th2, and Th17 cells can be categorized as non-Tfh Teff cells in parallel with Tfh and Treg cells, since both Tfh cells and Treg cells may be further divided into Th1-, Th2-, and Th17-like subpopulations [7][46][47][48]. Cell plasticity may exist between non-Tfh and Tfh cells, and between Teff cells and Treg cells [49][50][51]. However, even within Teff cells, particularly for Th17 cells, cell plasticity has been well documented. Th1-like Th17 cells have been found during infections and in autoimmune settings. The heterogeneity and plasticity of CD4 T cell subsets are largely determined by the temporal and quantitative co-expression of multiple master transcription factors during the differentiation of these cell subsets [7]. Between non-Tfh and Tfh cells, their balance may determine the magnitude of cellular versus humoral responses. In contrast, the balance between Teff and Treg cells reveals the yin and yang sides of the immune system. Understanding such balances is not only important for gaining our knowledge in basic immunology in infections and inflammations, but also for providing great opportunities in treating different types of immunological diseases. In fact, more and more attention is being paid to Treg-based therapies [52], because Treg cells play critical roles in anti-inflammation, autoimmunity prevention, and cancer promotion.
With the rapid development of modern technologies, such as single-cell RNA sequencing [53], CRISPR (clustered regularly interspaced short palindromic repeats)-mediated genome editing [54][55], Cryo-EM (cryogenic electron microscopy) structure analysis [56], single-cell mass spectrometry [57], and immunoimaging [58][59][60], many important findings are being made in the identification, development, and regulation of new CD4 T cell and ILC subsets, as well as their relationship and crosstalks in diseases. In the future, it is also important to investigate how CD4 T cell memory for each subset is formed and maintained [61] and whether the plasticity of antigen cross-reactive CD4 T cell memory cells may shape the primary response to a new infection [62]. With exciting new findings in basic immunology, our better and deeper understanding of this powerful system will help us uncover innovative measures for diseases treatment, thereby improving human health.
References
- Pennington, D.J.; Vermijlen, D.; Wise, E.L.; Clarke, S.L.; Tigelaar, R.E.; Hayday, A.C. The integration of conventional and unconventional T cells that characterizes cell-mediated responses. Adv. Immunol. 2005, 87, 27–59.
- Girardi, S.R.A.M. Conventional and Unconventional T Cells. Clin. Basic Immunodermatol. 2008, 6, 85–104.
- Kumar, B.V.; Connors, T.J.; Farber, D.L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213.
- Ruterbusch, M.; Pruner, K.B.; Shehata, L.; Pepper, M. In Vivo CD4(+) T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu. Rev. Immunol. 2020, 38, 705–725.
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010, 28, 445–489.
- Annunziato, F.; Romagnani, C.; Romagnani, S. The 3 major types of innate and adaptive cell-mediated effector immunity. J. Allergy Clin. Immunol. 2015, 135, 626–635.
- Fang, D.; Zhu, J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med. 2017, 214, 1861–1876.
- Chen, Y.; Kuchroo, V.K.; Inobe, J.; Hafler, D.A.; Weiner, H.L. Regulatory T cell clones induced by oral tolerance: Suppression of autoimmune encephalomyelitis. Science 1994, 265, 1237–1240.
- Groux, H.; O’Garra, A.; Bigler, M.; Rouleau, M.; Antonenko, S.; de Vries, J.E.; Roncarolo, M.G. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 1997, 389, 737–742.
- Dardalhon, V.; Awasthi, A.; Kwon, H.; Galileos, G.; Gao, W.; Sobel, R.A.; Mitsdoerffer, M.; Strom, T.B.; Elyaman, W.; Ho, I.C.; et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat. Immunol. 2008, 9, 1347–1355.
- Veldhoen, M.; Uyttenhove, C.; van Snick, J.; Helmby, H.; Westendorf, A.; Buer, J.; Martin, B.; Wilhelm, C.; Stockinger, B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 2008, 9, 1341–1346.
- Eyerich, S.; Eyerich, K.; Pennino, D.; Carbone, T.; Nasorri, F.; Pallotta, S.; Cianfarani, F.; Odorisio, T.; Traidl-Hoffmann, C.; Behrendt, H.; et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Investig. 2009, 119, 3573–3585.
- Devarajan, P.; Jones, M.C.; Kugler-Umana, O.; Vong, A.M.; Xia, J.; Swain, S.L. Pathogen Recognition by CD4 Effectors Drives Key Effector and Most Memory Cell Generation against Respiratory Virus. Front. Immunol. 2018, 9, 596.
- Juno, J.A.; van Bockel, D.; Kent, S.J.; Kelleher, A.D.; Zaunders, J.J.; Munier, C.M. Cytotoxic CD4 T Cells-Friend or Foe during Viral Infection? Front. Immunol. 2017, 8, 19.
- Takeuchi, A.; Saito, T. CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Front. Immunol. 2017, 8, 194.
- Eshima, K.; Chiba, S.; Suzuki, H.; Kokubo, K.; Kobayashi, H.; Iizuka, M.; Iwabuchi, K.; Shinohara, N. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4(+) Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol. Lett. 2012, 144, 7–15.
- Reis, B.S.; Hoytema van Konijnenburg, D.P.; Grivennikov, S.I.; Mucida, D. Transcription factor T-bet regulates intraepithelial lymphocyte functional maturation. Immunity 2014, 41, 244–256.
- Sledzinska, A.; Vila de Mucha, M.; Bergerhoff, K.; Hotblack, A.; Demane, D.F.; Ghorani, E.; Akarca, A.U.; Marzolini, M.A.V.; Solomon, I.; Vargas, F.A.; et al. Regulatory T Cells Restrain Interleukin-2- and Blimp-1-Dependent Acquisition of Cytotoxic Function by CD4(+) T Cells. Immunity 2020, 52, 151–166.e156.
- Cheroutre, H.; Husain, M.M. CD4 CTL: Living up to the challenge. Semin. Immunol. 2013, 25, 273–281.
- Mucida, D.; Husain, M.M.; Muroi, S.; van Wijk, F.; Shinnakasu, R.; Naoe, Y.; Reis, B.S.; Huang, Y.; Lambolez, F.; Docherty, M.; et al. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 2013, 14, 281–289.
- Reis, B.S.; Rogoz, A.; Costa-Pinto, F.A.; Taniuchi, I.; Mucida, D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat. Immunol. 2013, 14, 271–280.
- Serroukh, Y.; Gu-Trantien, C.; Hooshiar Kashani, B.; Defrance, M.; Vu Manh, T.P.; Azouz, A.; Detavernier, A.; Hoyois, A.; Das, J.; Bizet, M.; et al. The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. eLife 2018, 7, e30496.
- Donnarumma, T.; Young, G.R.; Merkenschlager, J.; Eksmond, U.; Bongard, N.; Nutt, S.L.; Boyer, C.; Dittmer, U.; Le-Trilling, V.T.; Trilling, M.; et al. Opposing Development of Cytotoxic and Follicular Helper CD4 T Cells Controlled by the TCF-1-Bcl6 Nexus. Cell Rep. 2016, 17, 1571–1583.
- Takeuchi, A.; Badr Mel, S.; Miyauchi, K.; Ishihara, C.; Onishi, R.; Guo, Z.; Sasaki, Y.; Ike, H.; Takumi, A.; Tsuji, N.M.; et al. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med. 2016, 213, 123–138.
- Marshall, N.B.; Vong, A.M.; Devarajan, P.; Brauner, M.D.; Kuang, Y.; Nayar, R.; Schutten, E.A.; Castonguay, C.H.; Berg, L.J.; Nutt, S.L.; et al. NKG2C/E Marks the Unique Cytotoxic CD4 T Cell Subset, ThCTL, Generated by Influenza Infection. J. Immunol. 2017, 198, 1142–1155.
- Meckiff, B.J.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Kusnadi, A.; Simon, H.; Eschweiler, S.; Grifoni, A.; Pelosi, E.; Weiskopf, D.; et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell 2020.
- Oh, D.Y.; Kwek, S.S.; Raju, S.S.; Li, T.; McCarthy, E.; Chow, E.; Aran, D.; Ilano, A.; Pai, C.S.; Rancan, C.; et al. Intratumoral CD4(+) T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 2020, 181, 1612–1625.
- Mattoo, H.; Stone, J.H.; Pillai, S. Clonally expanded cytotoxic CD4(+) T cells and the pathogenesis of IgG4-related disease. Autoimmunity 2017, 50, 19–24.
- Hashimoto, K.; Kouno, T.; Ikawa, T.; Hayatsu, N.; Miyajima, Y.; Yabukami, H.; Terooatea, T.; Sasaki, T.; Suzuki, T.; Valentine, M.; et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl. Acad. Sci. USA 2019, 116, 24242–24251.
- Gurram, R.K.; Zhu, J. Orchestration between ILC2s and Th2 cells in shaping type 2 immune responses. Cell. Mol. Immunol. 2019, 16, 225–235.
- Stockinger, B.; Omenetti, S. The dichotomous nature of T helper 17 cells. Nat. Rev. Immunol. 2017, 17, 535–544.
- Crotty, S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019, 50, 1132–1148.
- Miyauchi, K.; Sugimoto-Ishige, A.; Harada, Y.; Adachi, Y.; Usami, Y.; Kaji, T.; Inoue, K.; Hasegawa, H.; Watanabe, T.; Hijikata, A.; et al. Protective neutralizing influenza antibody response in the absence of T follicular helper cells. Nat. Immunol. 2016, 17, 1447–1458.
- Dominguez-Villar, M.; Hafler, D.A. Regulatory T cells in autoimmune disease. Nat. Immunol. 2018, 19, 665–673.
- Shevach, E.M. CD4+ CD25+ suppressor T cells: More questions than answers. Nat. Rev. Immunol. 2002, 2, 389–400.
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate Lymphoid Cells: 10 Years On. Cell 2018, 174, 1054–1066.
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301.
- Cherrier, D.E.; Serafini, N.; Di Santo, J.P. Innate Lymphoid Cell Development: A T Cell Perspective. Immunity 2018, 48, 1091–1103.
- Ebbo, M.; Crinier, A.; Vely, F.; Vivier, E. Innate lymphoid cells: Major players in inflammatory diseases. Nat. Rev. Immunol. 2017, 17, 665–678.
- Zhong, C.; Zheng, M.; Zhu, J. Lymphoid tissue inducer-A divergent member of the ILC family. Cytokine Growth Factor Rev. 2018, 42, 5–12.
- Sallusto, F. Heterogeneity of Human CD4(+) T Cells Against Microbes. Annu. Rev. Immunol. 2016, 34, 317–334.
- Hirahara, K.; Poholek, A.; Vahedi, G.; Laurence, A.; Kanno, Y.; Milner, J.D.; O’Shea, J.J. Mechanisms underlying helper T-cell plasticity: Implications for immune-mediated disease. J. Allergy Clin. Immunol. 2013, 131, 1276–1287.
- Leonard, W.J.; Lin, J.X.; O’Shea, J.J. The gammac Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity 2019, 50, 832–850.
- Strutt, T.M.; McKinstry, K.K.; Dibble, J.P.; Winchell, C.; Kuang, Y.; Curtis, J.D.; Huston, G.; Dutton, R.W.; Swain, S.L. Memory CD4+ T cells induce innate responses independently of pathogen. Nat. Med. 2010, 16, 558–564.
- Kawabe, T.; Jankovic, D.; Kawabe, S.; Huang, Y.; Lee, P.H.; Yamane, H.; Zhu, J.; Sher, A.; Germain, R.N.; Paul, W.E. Memory-phenotype CD4(+) T cells spontaneously generated under steady-state conditions exert innate TH1-like effector function. Sci. Immunol. 2017, 2.
- Fang, D.; Cui, K.; Mao, K.; Hu, G.; Li, R.; Zheng, M.; Riteau, N.; Reiner, S.L.; Sher, A.; Zhao, K.; et al. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J. Exp. Med. 2018, 215, 2705–2714.
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015, 349, 989–993.
- Yu, F.; Sharma, S.; Edwards, J.; Feigenbaum, L.; Zhu, J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat. Immunol. 2015, 16, 197–206.
- Zhu, J.; Paul, W.E. Heterogeneity and plasticity of T helper cells. Cell Res. 2010, 20, 4–12.
- Zhou, L.; Chong, M.M.; Littman, D.R. Plasticity of CD4+ T cell lineage differentiation. Immunity 2009, 30, 646–655.
- Sakaguchi, S.; Vignali, D.A.; Rudensky, A.Y.; Niec, R.E.; Waldmann, H. The plasticity and stability of regulatory T cells. Nat. Rev. Immunol. 2013, 13, 461–467.
- Raffin, C.; Vo, L.T.; Bluestone, J.A. Treg cell-based therapies: Challenges and perspectives. Nat. Rev. Immunol. 2020, 20, 158–172.
- Stubbington, M.J.T.; Rozenblatt-Rosen, O.; Regev, A.; Teichmann, S.A. Single-cell transcriptomics to explore the immune system in health and disease. Science 2017, 358, 58–63.
- Henriksson, J.; Chen, X.; Gomes, T.; Ullah, U.; Meyer, K.B.; Miragaia, R.; Duddy, G.; Pramanik, J.; Yusa, K.; Lahesmaa, R.; et al. Genome-wide CRISPR Screens in T Helper Cells Reveal Pervasive Crosstalk between Activation and Differentiation. Cell 2019, 176, 882–896.e818.
- Ledford, H. Quest to use CRISPR against disease gains ground. Nature 2020, 577, 156.
- Renaud, J.P.; Chari, A.; Ciferri, C.; Liu, W.T.; Remigy, H.W.; Stark, H.; Wiesmann, C. Cryo-EM in drug discovery: Achievements, limitations and prospects. Nat. Rev. Drug Discov. 2018, 17, 471–492.
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; El-ad, D.A.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696.
- Germain, R.N.; Robey, E.A.; Cahalan, M.D. A decade of imaging cellular motility and interaction dynamics in the immune system. Science 2012, 336, 1676–1681.
- Qi, H.; Kastenmuller, W.; Germain, R.N. Spatiotemporal basis of innate and adaptive immunity in secondary lymphoid tissue. Annu. Rev. Cell Dev. Biol. 2014, 30, 141–167.
- Tang, J.; van Panhuys, N.; Kastenmuller, W.; Germain, R.N. The future of immunoimaging—Deeper, bigger, more precise, and definitively more colorful. Eur. J. Immunol. 2013, 43, 1407–1412.
- Schreiner, D.; King, C.G. CD4+ Memory T Cells at Home in the Tissue: Mechanisms for Health and Disease. Front. Immunol. 2018, 9, 2394.
- Jameson, S.C.; Masopust, D. Understanding Subset Diversity in T Cell Memory. Immunity 2018, 48, 214–226.





