
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Daniel Hatlem | -- | 2901 | 2022-10-18 12:20:20 | | | |
| 2 | Peter Tang | -4 word(s) | 2897 | 2022-10-19 03:00:56 | | |
Video Upload Options
The SpyCatcher-SpyTag system was developed as a method for protein ligation. It is based on a modified domain from a Streptococcus pyogenes surface protein (SpyCatcher), which recognizes a cognate 13-amino-acid peptide (SpyTag). Upon recognition, the two form a covalent isopeptide bond between the side chains of a lysine in SpyCatcher and an aspartate in SpyTag. This technology has been used, among other applications, to create covalently stabilized multi-protein complexes, for modular vaccine production, and to label proteins (e.g., for microscopy).
1. Introduction
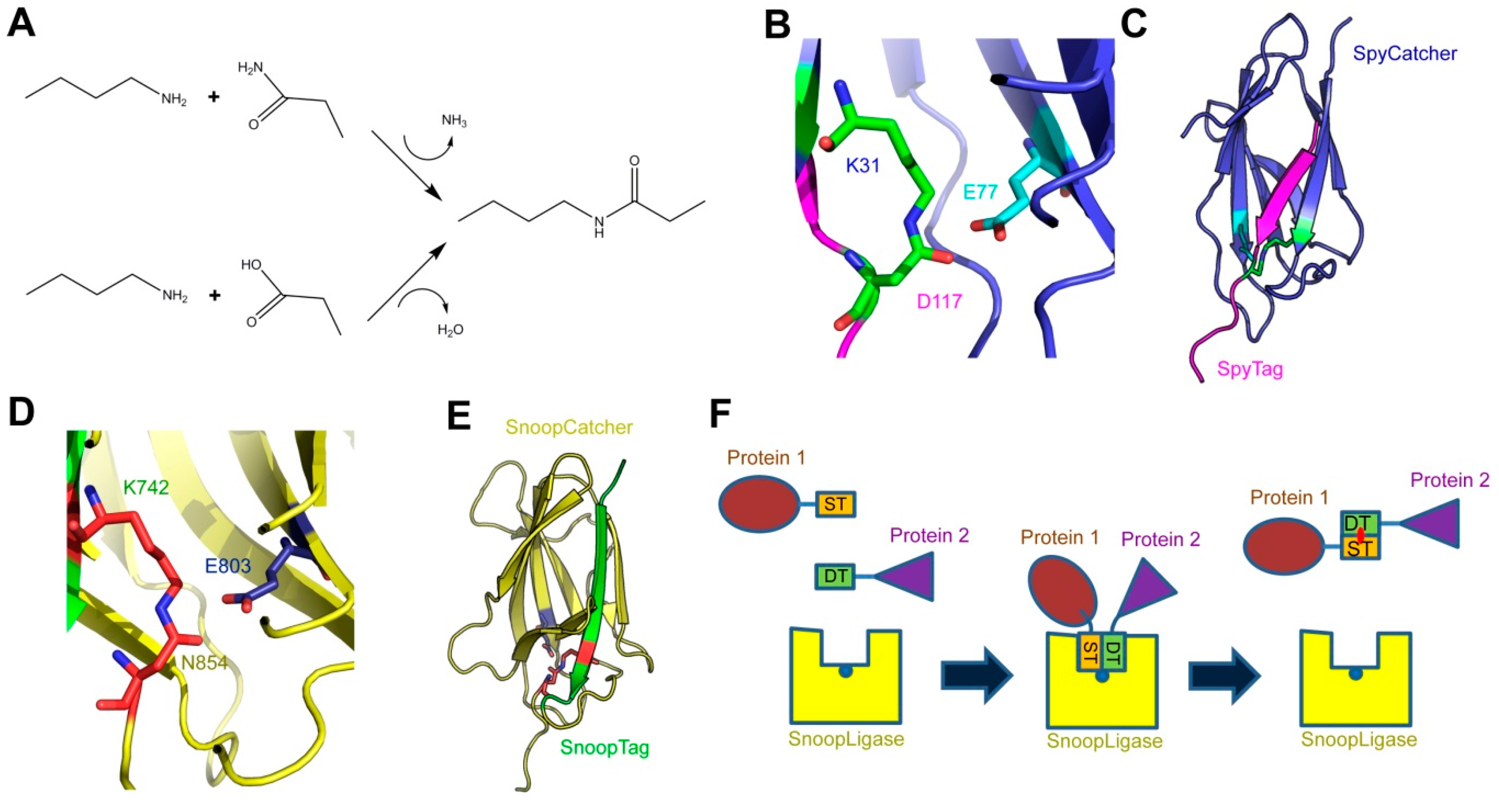
|
Catcher |
Tag |
Tag Sequence |
Description |
Publication Year |
Reference |
|---|---|---|---|---|---|
|
SpyCatcher |
SpyTag |
AHIVMVDAYKPTK 1 |
Original Catcher-Tag technology. |
2012 |
[11] |
|
SpyCatcher ΔN1ΔC1 |
SpyTag |
AHIVMVDAYKPTK 1 |
Minimal SpyCatcher construct that still binds efficiently to SpyTag. |
2014 |
[2] |
|
SpyLigase 2 |
SpyTag |
AHIVMVDAYKPTK 1 |
Rationally engineered system for ligating two peptides. |
2014 |
[17] |
|
KTag |
ATHIKFSKRD |
||||
|
SnoopCatcher |
SnoopTag |
KLGDIEFIKVNK 1 |
Orthogonal technology to SpyCatcher. |
2016 |
[18] |
|
SpyCatcher002 |
SpyTag002 |
VPTIVMVDAYKRYK 1 |
Improved SpyCatcher-SpyTag system with faster reaction rate. |
2017 |
[19] |
|
SnoopLigase 2 |
SnoopTagJr |
KLGSIEFIKVNK 1 |
Rationally engineered system for ligating two peptides. |
2018 |
[20] |
|
DogTag |
DIPATYEFTDGKHYITNEPIPPK |
||||
|
SpyDock |
SpyTag002 |
VPTIVMVDAYKRYK 1 |
Protein affinity purification system (Spy&Go) based on SpyCatcher. |
2019 |
[21] |
2. Applications of the SpyCatcher-SpyTag System

3. Using SpyCatcher-SpyTag to Investigate Bacterial Virulence Factors
References
- Koglin, A.; Walsh, C.T. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 2009, 26, 987–1000.
- Li, L.; Fierer, J.O.; Rapoport, T.A.; Howarth, M. Structural analysis and optimization of the covalent association between SpyCatcher and a peptide Tag. J. Mol. Biol. 2014, 426, 309–317.
- Izoré, T.; Contreras-Martel, C.; El Mortaji, L.; Manzano, C.; Terrasse, R.; Vernet, T.; Di Guilmi, A.M.; Dessen, A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 2010, 18, 106–115.
- Collins, G.A.; Goldberg, A.L. The logic of the 26S proteasome. Cell 2017, 169, 792–806.
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157.
- Wilson, V.G. Introduction to Sumoylation. Adv. Exp. Med. Biol. 2017, 963, 1–12.
- Baker, E.N.; Squire, C.J.; Young, P.G. Self-generated covalent cross-links in the cell-surface adhesins of Gram-positive bacteria. Biochem. Soc. Trans. 2015, 43, 787–794.
- Hendrickx, A.P.; Budzik, J.M.; Oh, S.-Y.; Schneewind, O. Architects at the bacterial surface—Sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 2011, 9, 166–176.
- Sridharan, U.; Ponnuraj, K. Isopeptide bond in collagen- and fibrinogen-binding MSCRAMMs. Biophys. Rev. 2016, 8, 75–83.
- Terao, Y.; Kawabata, S.; Nakata, M.; Nakagawa, I.; Hamada, S. Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 2002, 277, 47428–47435.
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697.
- Hagan, R.M.; Björnsson, R.; McMahon, S.A.; Schomburg, B.; Braithwaite, V.; Bühl, M.; Naismith, J.H.; Schwarz-Linek, U. NMR spectroscopic and theoretical analysis of a spontaneously formed Lys-Asp isopeptide bond. Angew. Chem. Int. Ed. Engl. 2010, 49, 8421–8425.
- Zhang, W.-B.; Sun, F.; Tirrell, D.A.; Arnold, F.H. Controlling macromolecular topology with genetically encoded SpyTag-SpyCatcher chemistry. J. Am. Chem. Soc. 2013, 135, 13988–13997.
- Bedbrook, C.N.; Kato, M.; Kumar, S.R.; Lakshmanan, A.; Nath, R.D.; Sun, F.; Sternberg, P.W.; Arnold, F.H.; Gradinaru, V. Genetically encoded Spy peptide fusion system to detect plasma membrane-localized proteins in vivo. Chem. Biol. 2015, 22, 1108–1121.
- Chauhan, N.; Hatlem, D.; Orwick-Rydmark, M.; Schneider, K.; Floetenmeyer, M.; van Rossum, B.; Leo, J.C.; Linke, D. Insights into the autotransport process of a trimeric autotransporter, Yersinia Adhesin A (YadA). Mol. Microbiol. 2019, 111, 844–862.
- Brune, K.D.; Howarth, M. New routes and opportunities for modular construction of particulate vaccines: Stick, click, and glue. Front. Immunol. 2018, 9, 1432.
- Fierer, J.O.; Veggiani, G.; Howarth, M. SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc. Natl. Acad. Sci. USA 2014, 111, E1176–E1181.
- Veggiani, G.; Nakamura, T.; Brenner, M.D.; Gayet, R.V.; Yan, J.; Robinson, C.V.; Howarth, M. Programmable polyproteams built using twin peptide superglues. Proc. Natl. Acad. Sci. USA 2016, 113, 1202–1207.
- Keeble, A.H.; Banerjee, A.; Ferla, M.P.; Reddington, S.C.; Anuar, I.N.; Howarth, M. Evolving Accelerated Amidation by SpyTag/SpyCatcher to Analyze Membrane Dynamics. Angew. Chem. Int. Ed. Engl. 2017, 56, 16521–16525.
- Buldun, C.M.; Jean, J.X.; Bedford, M.R.; Howarth, M. SnoopLigase catalyzes peptide-peptide locking and enables solid-phase conjugate isolation. J. Am. Chem. Soc. 2018, 140, 3008–3018.
- Anuar, I.N.; Banerjee, A.; Keeble, A.H.; Carella, A.; Nikov, G.I.; Howarth, M. Spy&Go purification of SpyTag-proteins using pseudo-SpyCatcher to access an oligomerization toolbox. Nat. Commun. 2019, 10, 1743.
- Veggiani, G.; Zakeri, B.; Howarth, M. Superglue from bacteria: Unbreakable bridges for protein nanotechnology. Trends Biotechnol. 2014, 32, 506–512.
- Reddington, S.C.; Howarth, M. Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher. Curr. Opin. Chem. Biol. 2015, 29, 94–99.
- Keeble, A.H.; Howarth, M. Insider information on successful covalent protein coupling with help from SpyBank. Meth. Enzym. 2019, 617, 443–461.
- Si, M.; Xu, Q.; Jiang, L.; Huang, H. SpyTag/SpyCatcher cyclization enhances the thermostability of firefly luciferase. PLoS ONE 2016, 11, e0162318.
- Schoene, C.; Fierer, J.O.; Bennett, S.P.; Howarth, M. SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme. Angew. Chem. Int. Ed. Engl. 2014, 53, 6101–6104.
- Sun, X.-B.; Cao, J.-W.; Wang, J.-K.; Lin, H.-Z.; Gao, D.-Y.; Qian, G.-Y.; Park, Y.-D.; Chen, Z.-F.; Wang, Q. SpyTag/SpyCatcher molecular cyclization confers protein stability and resilience to aggregation. New Biotechnol. 2018, 49, 28–36.
- Schoene, C.; Bennett, S.P.; Howarth, M. SpyRing interrogation: Analyzing how enzyme resilience can be achieved with phytase and distinct cyclization chemistries. Sci. Rep. 2016, 6, 21151.
- Wang, J.; Wang, Y.; Wang, X.; Zhang, D.; Wu, S.; Zhang, G. Enhanced thermal stability of lichenase from Bacillus subtilis 168 by SpyTag/SpyCatcher-mediated spontaneous cyclization. Biotechnol. Biofuels 2016, 9, 79.
- Schoene, C.; Bennett, S.P.; Howarth, M. SpyRings declassified: A blueprint for using isopeptide-mediated cyclization to enhance enzyme thermal resilience. Methods Enzym. 2016, 580, 149–167.
- Langer, R.; Tirrell, D.A. Designing materials for biology and medicine. Nature 2004, 428, 487–492.
- Guilak, F.; Butler, D.L.; Goldstein, S.A.; Baaijens, F.P.T. Biomechanics and mechanobiology in functional tissue engineering. J. Biomech. 2014, 47, 1933–1940.
- Lee, K.Y.; Mooney, D.J. Hydrogels for tissue engineering. Chem. Rev. 2001, 101, 1869–1879.
- Caliari, S.R.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414.
- Burdick, J.A.; Murphy, W.L. Moving from static to dynamic complexity in hydrogel design. Nat. Commun. 2012, 3, 1269.
- Gao, X.; Fang, J.; Fu, L.; Li, H. Engineering protein hydrogels using SpyCatcher-SpyTag chemistry. Biomacromolecules 2016, 17, 2812–2819.
- Wang, R.; Yang, Z.; Luo, J.; Hsing, I.-M.; Sun, F. B12-dependent photoresponsive protein hydrogels for controlled stem cell/protein release. Proc. Natl. Acad. Sci. USA 2017, 114, 5912–5917.
- Gao, X.; Lyu, S.; Li, H. Decorating a blank slate protein hydrogel: A general and robust approach for functionalizing protein hydrogels. Biomacromolecules 2017, 18, 3726–3732.
- Liu, X.; Yang, X.; Yang, Z.; Luo, J.; Tian, X.; Liu, K. Versatile engineered protein hydrogels enabling decoupled mechanical and biochemical tuning for cell adhesion and neurite growth. ACS Appl. Nano Mater. 2018, 1, 1579–1585.
- Ma, W.; Saccardo, A.; Roccatano, D.; Aboagye-Mensah, D.; Alkaseem, M.; Jewkes, M.; Di Nezza, F.; Baron, M.; Soloviev, M.; Ferrari, E. Modular assembly of proteins on nanoparticles. Nat. Commun. 2018, 9, 1489.
- Alves, N.J.; Turner, K.B.; Daniele, M.A.; Oh, E.; Medintz, I.L.; Walper, S.A. Bacterial nanobioreactors—Directing enzyme packaging into bacterial outer membrane vesicles. ACS Appl. Mater. Interfaces 2015, 7, 24963–24972.
- Giessen, T.W.; Silver, P.A. A catalytic nanoreactor based on in vivo encapsulation of multiple enzymes in an engineered protein nanocompartment. ChemBioChem 2016, 17, 1931–1935.
- Thrane, S.; Janitzek, C.M.; Matondo, S.; Resende, M.; Gustavsson, T.; de Jongh, W.A.; Clemmensen, S.; Roeffen, W.; van de Vegte-Bolmer, M.; van Gemert, G.J.; et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines. J. Nanobiotechnol. 2016, 14, 30.
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234.
- Zhao, Y.; Zhang, Y.; Teng, Y.; Liu, K.; Liu, Y.; Li, W.; Wu, L. SpyCLIP: An easy-to-use and high-throughput compatible CLIP platform for the characterization of protein-RNA interactions with high accuracy. Nucleic Acids Res. 2019, 6, 3.
- Maticzka, D.; Ilik, I.A.; Aktas, T.; Backofen, R.; Akhtar, A. uvCLAP is a fast and non-radioactive method to identify in vivo targets of RNA-binding proteins. Nat. Commun. 2018, 9, 1142.
- Gu, J.; Wang, M.; Yang, Y.; Qiu, D.; Zhang, Y.; Ma, J.; Zhou, Y.; Hannon, G.J.; Yu, Y. GoldCLIP: Gel-omitted Ligation-dependent CLIP. Genom. Proteom. Bioinform. 2018, 16, 136–143.
- England, C.G.; Luo, H.; Cai, W. HaloTag technology: A versatile platform for biomedical applications. Bioconjugate Chem. 2015, 26, 975–986.
- Moon, H.; Bae, Y.; Kim, H.; Kang, S. Plug-and-playable fluorescent cell imaging modular toolkits using the bacterial superglue, SpyTag/SpyCatcher. Chem. Commun. 2016, 52, 14051–14054.
- Hinrichsen, M.; Lenz, M.; Edwards, J.M.; Miller, O.K.; Mochrie, S.G.; Swain, P.S.; Schwarz-Linek, U.; Regan, L. A new method for post-translationally labeling proteins in live cells for fluorescence imaging and tracking. Protein Eng. Des. Sel. 2017, 30, 771–780.
- Pessino, V.; Citron, Y.R.; Feng, S.; Huang, B. Covalent protein labeling by SpyTag-SpyCatcher in fixed cells for super-resolution microscopy. ChemBioChem 2017, 18, 1492–1495.
- Grenier, V.; Daws, B.R.; Liu, P.; Miller, E.W. Spying on neuronal membrane potential with genetically targetable voltage indicators. J. Am. Chem. Soc. 2019, 141, 1349–1358.
- Yin, L.; Guo, X.; Liu, L.; Zhang, Y.; Feng, Y. Self-assembled multimeric-enzyme nanoreactor for robust and efficient biocatalysis. ACS Biomater. Sci. Eng. 2018, 4, 2095–2099.
- Fan, E.; Chauhan, N.; Udatha, D.B.; Leo, J.C.; Linke, D. Type V Secretion Systems in Bacteria. Microbiol. Spectr. 2016, 4.
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359.
- Chagnot, C.; Zorgani, M.A.; Astruc, T.; Desvaux, M. Proteinaceous determinants of surface colonization in bacteria: Bacterial adhesion and biofilm formation from a protein secretion perspective. Front. Microbiol. 2013, 4, 303.
- Feilmeier, B.J.; Iseminger, G.; Schroeder, D.; Webber, H.; Phillips, G.J. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000, 182, 4068–4076.
- Meiresonne, N.Y.; Consoli, E.; Mertens, L.M.Y.; Chertkova, A.O.; Goedhart, J.; den Blaauwen, T. Superfolder mTurquoise2ox optimized for the bacterial periplasm allows high efficiency in vivo FRET of cell division antibiotic targets. Mol. Microbiol. 2019, 111, 1025–1038.
- Dinh, T.; Bernhardt, T.G. Using superfolder green fluorescent protein for periplasmic protein localization studies. J. Bacteriol. 2011, 193, 4984–4987.
- El Khatib, M.; Martins, A.; Bourgeois, D.; Colletier, J.-P.; Adam, V. Rational design of ultrastable and reversibly photoswitchable fluorescent proteins for super-resolution imaging of the bacterial periplasm. Sci. Rep. 2016, 6, 18459.
- Meiresonne, N.Y.; van der Ploeg, R.; Hink, M.A.; den Blaauwen, T. Activity-Related Conformational Changes in d,d-Carboxypeptidases Revealed by In Vivo Periplasmic Förster Resonance Energy Transfer Assay in Escherichia coli. MBio 2017, 8, e01089-17.
- Rassam, P.; Copeland, N.A.; Birkholz, O.; Tóth, C.; Chavent, M.; Duncan, A.L.; Cross, S.J.; Housden, N.G.; Kaminska, R.; Seger, U.; et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature 2015, 523, 333–336.
- Toseland, C.P. Fluorescent labeling and modification of proteins. J. Chem. Biol. 2013, 6, 85–95.
- Morales, A.R.; Schafer-Hales, K.J.; Marcus, A.I.; Belfield, K.D. Amine-reactive fluorene probes: Synthesis, optical characterization, bioconjugation, and two-photon fluorescence imaging. Bioconjugate Chem. 2008, 19, 2559–2567.
- Jose, J.; von Schwichow, S. “Cystope tagging” for labeling and detection of recombinant protein expression. Anal. Biochem. 2004, 331, 267–274.
- Zhou, Z.; Cironi, P.; Lin, A.J.; Xu, Y.; Hrvatin, S.; Golan, D.E.; Silver, P.A.; Walsh, C.T.; Yin, J. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem. Biol. 2007, 2, 337–346.
- Zhang, G.; Zheng, S.; Liu, H.; Chen, P.R. Illuminating biological processes through site-specific protein labeling. Chem. Soc. Rev. 2015, 44, 3405–3417.





