
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | 佳颖 李 | -- | 2683 | 2022-10-11 15:15:01 | | | |
| 2 | Sirius Huang | -1 word(s) | 2682 | 2022-10-14 08:58:31 | | |
Video Upload Options
1. Introduction
Colorectal cancer (CRC) ranks third in the incidence of malignant tumors and is the second leading cause of cancer-related death worldwide [1][2]. According to the global cancer statistics issued by International Agency for Research on Cancer (IARC), 1.9 million new cases (third highest incidence) and 935,000 colorectal cancer deaths (second highest mortality) were estimated to have occurred worldwide in 2020 [3] . The global burden of colorectal cancer is expected to be more than 2.2 million new cases a year and 1.1 million deaths in 2030 [4]. In China, the incidence and mortality of colorectal cancer have also increased due to variations in diet and the population age structure [5][6]. Due to the lack of early specific warning signs of colorectal cancer, most patients are in phases III and IV at the first visit and might lose the opportunity to receive effective standard treatment, resulting in a 5-year survival rate of 40% [7][8]. Therefore, it is urgent to find new therapeutic methods and develop effective biomarkers for the early diagnosis, treatment, and prognosis assessment of colorectal cancer to improve the survival status. Some studies have demonstrated that epigenetic mechanisms play a key role in cancer progression, particularly non-coding RNAs (ncRNAs) [9]. Indeed, many studies have demonstrated that the development pathogenesis of colorectal cancer is highly influenced by ncRNAs [10], the abnormal expression of oncogenic and tumor-suppressor molecules, and the abnormal activation of various cell signaling pathways [11][12]. In recent years, some important natural compounds, such as phenolics, terpenoids, and meroterpenoids, have been confirmed to have anticancer effects by regulating the expression and function of ncRNAs [13]. Particularly, naturally derived polyphenols have been safely used for many years and have shown real potential for therapeutic effects in most cancers through the regulation of miRNAs and lncRNAs [14][15][16]. The initial epigenetic changes associated with cancer may be regulated by many polyphenols [17][18][19], such as curcumin [20][21], resveratrol [22][23], and so on. Among them, curcumin, with the advantages of less toxicity and less side effects, has been clarified to be an effective compound in the treatment of colorectal cancer. Therefore, summarizing the scientific progress of curcumin targeting ncRNAs in colorectal cancer and understanding the inducement and molecular regulatory mechanisms of colorectal cancer is essential in finding key targets for clinical treatment and will also provide theoretical guidance for basic research, clinical drug selection, and gene therapy.
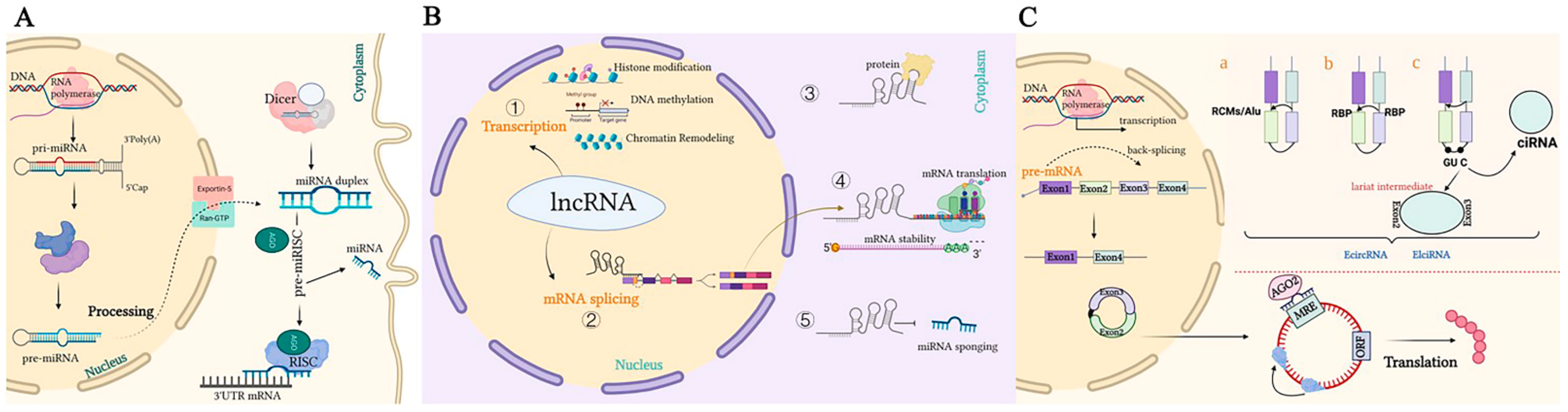
Figure 1. Representation of the biogenesis and mode of action of ncRNAs. (A). A pri-miRNA with a double-stranded stem-loop is formed after transcription, which is cleaved by Drosha and DGCR8 with a hairpin-based secondary structure; two nucleotides overhang at its 3′ end. Then, pre-miRNAs are exported to the cytoplasm by exportin-5/Ran-GTP. Here, pre-miRNA forms a mature double-stranded miRNA duplex digested by Dicer. The miRISC complex formed by the guide strand miRNA and Ago protein represses target mRNAs by base-pairing at 3‘UTR, which prevents translation and selectively silences gene expression. (B). Mechanisms underlying long non-coding RNA (lncRNA)-mediated regulation of gene expression. ① Transcription regulation by lncRNAs. ② lncRNAs are engaged in the processing and maturation of mRNAs ③ lncRNAs interact with proteins. ④ lncRNAs interact with RNAs. ⑤ lncRNAs can competitively bind to miRNAs by acting as ceRNAs, thereby blocking the inhibition of the target gene. (C). Biogenesis of circRNAs. a. The back-splicing circularization requires the help of complementary sequences (ALU repeats and RCMs). b. RBP-mediated circularization. c. Lariat-driven circularization. circRNA can serve as a miRNA sponge, which inhibits miRNAs in order to regulate the expression of target genes or interact with proteins.
2. Curcumin and Colorectal Cancer Therapy Based on Non-Coding RNAs’ Epigenetic Regulation
2.1. Curcumin Against Colorectal Cancer Mediated by miRNAs
Many studies have demonstrated that the occurrence, development, treatment, and prognosis of colorectal cancer are all involved with miRNAs in varying degrees [24][25]. miRNAs can regulate the expression of their target oncogenes or cancer suppressor genes to affect the various biological processes of cancer, including cell proliferation, apoptosis, cell cycle, and metastases, in the occurrence and development of malignant tumors [26]. Some studies have indicated that curcumin could exert anti-colorectal cancer effects by targeting differentially expressed miRNAs [27][28]. The miRNAs regulated by curcumin in colorectal cancer are summarized in Table 1.
Table 1. Curcumin modulates miRNAs in colorectal cancer.
|
In Vitro/ In Vivo |
Cell Line |
Modulated by Curcumin |
Target Gene |
Relevant Mechanism |
Biological Effects After Administration |
Refs |
|
In vitro/ in vivo |
Rko, HCT116, HT-29, SW620 |
miR-21 ↓ miR-21-3p, miR-21-5p ↓ |
PDCD4 PTEN ATG10, APAF1 |
Suppress AP-1 binding to the promoter, p-Akt |
Inhibits tumor growth, migration, invasion, metastasis. Promotes autophagy, apoptosis |
[29] [30]
|
|
In vitro |
SW480 |
miR-130a ↓ |
— |
Wnt/β-catenin pathway |
Inhibits proliferation |
[31] |
|
In vitro/ in vivo |
HCT116, SW480, HCT116p53−/− |
miR-34a ↑ miR-27a ↓ |
CDK4, CDK6, cMyc, FBXW7, Cyclin D1 |
Deregulation of miRNAs — |
Inhibits proliferation, tumor growth, chemoresistant. Promotes cell cycle arrest, apoptosis |
[32] |
|
In vitro |
HCT-116 |
miR-491 ↑ |
PEG10 |
Wnt/β-catenin pathway |
Inhibits proliferation and promotes apoptosis |
[33] |
|
In vitro |
HCT116−5FUR, SW480-5FUR |
miR-141, miR-101, miR-200b, miR-429, miR-200c ↑ |
— ZEB1 BMI1 |
— |
Inhibits EMT |
[34] |
|
In vitro |
HCT-116, L-OHP |
miR-409-3p ↑ |
ERCC1 |
— |
Inhibits migration and invasion; promotes apoptosis. |
[35] |
|
In vitro |
RKO, SW480 |
miR-20a, miR-27a, miR-17 ↓ |
ZBTB4, ZBTB10, Sp1, Sp3, Sp4, |
— |
Inhibits proliferation |
[36] |
|
In vitro/in vivo |
SW620, HCT116, HCT116wt, HCT116 p53−/− |
miR-34a ↑ miR-34c ↑ |
Notch-1 |
— |
Inhibits proliferation, promotes apoptosis |
[37] |
|
In vitro/in vivo |
SW480 |
miR-145 ↑ (Nano-CUR) |
— |
— |
Interferes with tumor growth, inhibits proliferation, migration |
[38] |
|
In vitro/in vivo |
HCT116, LoVo, HT29-MTX |
miR-31 (PS-TP-miR-31i/Cur NPs) |
— |
— |
Inhibits cell proliferation, tumor growth |
[39] |
|
In vitro |
HT-29, HCT-116, LoVo, SW480, DLD-1 CRL-1790 |
miR-137 ↓ |
GLS |
GLS–gluamine Metabolism |
Increases cell death, anti-chemoresistance |
[40] |
|
In vitro |
HT-29 |
miR-21, miR-155, miR221/222 ↓ miR-34a, miR-126 ↑ |
— |
— |
Promotes apoptosis |
[41] |
Note: Arrows “↓, ↑” represent the expression levels of miRNAs regulated by curcumin in CRC. “↑”: upregulate “↓” :downregulate
2.2. Curcumin against Colorectal Cancer Mediated by LncRNAs
LncRNAs, as effective marker molecules, have been used in the diagnosis and prognosis of many cancers. The abnormal expression of some lncRNAs is involved in various processes, such as regulating the growth and metastasis of tumor cells [42]. Curcumin could significantly change the proliferation, migration, and invasion of colorectal cancer cells by regulating lncRNAs [43]. Furthermore, the mechanism of lncRNAs that interact with curcumin to moderate signal transduction needs to be further explored. Therefore, this section will discuss the intervention effect between curcumin and lncRNAs in colorectal cancer and its anti-colorectal cancer mechanism. The above-mentioned results have gradually provided new evidence for the basic research of lncRNAs combined with curcumin for the treatment of colorectal cancer. The data are shown in Table 2.
Table 2. Curcumin modulates lncRNAs in colorectal cancer.
|
In Vitro/ In Vivo |
Cell Line |
Modulated by Curcumin |
Relevant Mechanism |
Biological Effects after Administration |
Refs |
|---|---|---|---|---|---|
|
In vitro |
HCT116, SW480 |
NBR2 ↑ |
Activates of AMPK pathway |
Inhibits proliferation |
[44] |
|
In vitro/ In vivo |
HCT8 DDP cells |
KCNQ1OT1 ↓ |
Sponge of miR-497 increases Bcl-2 expression |
Inhibits proliferation, promotes apoptosis, chemoresistant |
[45] |
|
In vitro |
DLD-1, SW620, HCT116 |
PANDAR ↓ |
PUMA upregulation |
Promotes apoptosis |
[46] |
|
In vitro/ In vivo |
DLD-1 |
PANDAR ↓ |
Induces PUMA |
Promotes apoptosis, reduces cell aging |
|
|
In vitro |
SW480 |
MALAT1 ↓ (Si-MALAT1) |
Downregulated c-myc, cyclinD1, β-catenin |
Inhibits cell viability, migration, invasion |
[47] |
|
In vitro/ In vivo |
HT-29 |
CCAT1 ↓ (Si-CCAT1-CSNP) |
— |
Inhibits proliferation, migration, induces apoptosis |
[48] |
Note: Arrows “↓, ↑” represent the expression levels of lncRNAs regulated by curcumin in CRC. “↑ : upregulate “↓” :downregulate
2.3. Curcumin and Anti-tumor Effect Mediated by CircRNAs
circRNAs have great potential research value in the occurrence, development, diagnosis, prognosis, and treatment of tumors. Recent studies have revealed that phytochemicals such as curcumin could further exert anti-tumor effects by regulating circRNAs that are engaged in biological processes, including tumor cell proliferation, apoptosis, migration, invasion, autophagy, chemosensitivity, and radiosensitivity [49]. Considering the pivotal roles of circRNAs combined with drugs in cancer, researchers found that curcumin could regulate the occurrence and development of various tumors through circRNAs acting on different signaling pathways. The research results of the role of circRNAs in various tumors under the action of curcumin in the past 12 years are summarized in Table 3.
Table 3. Curcumin modulates circRNAs in various cancers.
|
In Vitro/ In Vivo |
Cell Line |
Modulated by Curcumin |
Relevant Mechanism |
Biological Effects after Administration |
Refs |
|
In vitro/ In vivo |
H1650, H1299, H460, A549, 16HBE (NSCLC) |
circ-PRKCA ↓ |
circ-PRKCA/miR-384/ITGB1 pathway |
Inhibits viability, colony formation, migration, invasion, promotes apoptosis |
[50] |
|
In vitro/ In vivo |
THLE-2, HuH-7, HCCLM3 (HCC) |
circ-ZNF83 ↓ |
JAK2/STAT3 pathway circZNF83/miR-324-5p/CDK16 axis |
Inhibits proliferation, cell cycle, migration, invasion, promotes apoptosis |
[51] |
|
In vitro/ In vivo |
SKOV3, A2780, IOSE-80 (Ovarian cancer) |
circ-PLEKHM3 ↑ |
circ-PLEKHM3/miR-320a/SMG1 axis |
Inhibits proliferation and promotes apoptosis |
[52] |
|
In vitro/ In vivo |
CAKI-1, ACHN, (RCC) |
circ-FNDC3B ↓ |
circ-FNDC3B/miR-138-5p/IGF2 axis |
Inhibits proliferation, promotes apoptosis |
[53] |
|
In vitro |
CNE-2 (NPC) |
circRNA-102115 |
circRNA-102115/miR-335-3p/MAPK1 |
Enhances radiosensitization |
[54] |
|
In vitro |
CNE-2 (NPC) |
circRNA network |
miRNA sponge regulated EGFR, STAT3, GRB2 |
Enhances radiosensitization |
[55] [56] |
Note: Arrows “↓, ↑” represent the expression levels of circRNAs regulated by curcumin in various cancers. “↑”: upregulate “↓” :downregulate
Meanwhile, researchers found evidence for the potential role of some circRNAs as diagnostic markers for colorectal cancer, as shown in Table 4.
Table 4. Roles of circRNAs in colorectal cancer.
|
In Vitro/In Vivo |
Cell Line |
circRNAs in CRC |
Target Gene |
Relevant Mechanism |
Biological Effects |
Refs |
|
In vitro/ In vivo |
RKO, HCT116, SW480, SW620, DLD-1, HT29, Colo320, HCE8693 |
circ-RHOBTB3 ↓ |
PTBP1, FUS ADARB2 |
circ-RHOBTB3/HuR/PTBP1 protein ubiquitination |
Restrains metastasis, invasion |
[57] |
|
In vitro/ In vivo |
HCT116, SW480 |
circ-3823 ↑ |
TCF7 |
circ-3823/miR-30c-5p/TCF7 miRNA sponge m6A modification |
Promotes proliferation, metastasis, angiogenesis |
[58] |
|
In vitro |
FHC, HCT116, DLD1, LoVo, SW620, HT29, SW480 |
circ-IL4R ↑ |
PHLPP1 |
circ-IL4R/PI3K/AKT, miRNA sponge, protein ubiquitination |
Promotes proliferation, migration, invasion |
[59] |
|
In vitro/ In vivo |
HT-29, SW480, HCT-116, LoVo, NCM460 |
circ-N4BP2L2 ↑ |
CXCR4 |
circ-N4BP2L2/miR-340-5p/ CXCR4 (miRNA sponge) |
Promotes proliferation, migration, invasion. Promotes tumor growth, metastasis |
[60] |
|
In vitro/ In vivo |
HT290, HCT116, SW480, SW620, FHC |
circ-CUL2 ↓ |
PPP6C |
circ-CUL2/miR-208a-3p/PPP6C (miRNA sponge) |
Inhibits proliferation ability, induces apoptosis, autophagy |
[61] |
|
In vitro/ In vivo |
HT-29, LoVo, SW480, HCT-116, NCM460 |
circ-PTK2 ↑ |
YTHDF1 |
circ-PTK2/miR-136-5p/YTHDF1 |
Promotes proliferation, migration, invasion, chemoresistance |
[62] |
Note: Arrows “↓, ↑” represent the expression levels of circRNAs in CRC. “↑ : upregulate “↓” :downregulate
Throughout the last decade of research, researchers have identified a series of circRNAs [63] as biomarkers of oncogenic/tumor-suppressive genes that exert anti-colorectal-cancer functions. Although the regulation of these functional molecules of circRNAs has gradually entered public view, the research on circRNAs has just begun. These circRNAs may provide new evidence for future research on the mechanism of colorectal cancer occurrence and development. Whether those circRNAs are responsible for the treatment of colorectal cancer and how circRNAs regulate the biological effects of tumor cells after the curcumin intervention have still not been studied. There is still plenty of room to explore the effect of curcumin on colorectal cancer through circRNAs.
3. Discussion
Curcumin can interact with multiple molecular targets through a variety of complex molecular mechanisms to inhibit the growth of tumor cells and achieve anti-colorectal-cancer and chemotherapy sensitization effects. Compared with traditional chemotherapy drugs, curcumin has higher safety levels and is widely used as an ingredient in dietary formulations for the prevention of colorectal cancer. Related clinical trials have been launched. The evidence listed above identifies the mechanism of ncRNAs as potential targets of curcumin for colorectal cancer treatment, summarized in Figure 2. As an explicit regulatory mechanism, miRNAs regulate target mRNAs through complete or incomplete base pairing with 3′UTRs (Figure 2-ⅰ), lncRNAs interact with DNAs, RNAs, and proteins to regulate transcription and post-transcriptional processes (Figure 2-ⅱ); circRNAs interact with proteins to regulate the alternative splicing and transcription of target genes (Figure 2-ⅲ). In particular, current studies on ncRNAs regulated by curcumin have been mainly focused on the ceRNA mechanism, also known as the molecular sponge effect, which mainly involves the lncRNAs/circRNAs–miRNAs–mRNAs–proteins pathway. Briefly, target genes can be silenced by miRNA binding. However, ceRNAs, including lncRNAs and circRNAs, can regulate target gene expression by competitively absorbing miRNAs. The mutual regulation between these transcripts (mRNAs and ncRNAs) plays an important role in the occurrence and development of colorectal cancer and mediates biological processes, including colorectal cancer cell proliferation, apoptosis, metastasis, and chemoresistance (Figure 2-ⅳ). As a new research tool or idea, ceRNAs have also been crossed, penetrated, and merged with research in drug-related fields, such as in research on curcumin. Researchers found that curcumin has shown a significant anti-tumor effect in the epigenetic regulation of ncRNAs according to the research progress in the past 12 years. Curcumin could affect the development of colorectal cancer by targeting oncogenes such as miR-130a/miR-137, miR-20a/miR-27a, miR-21, and miR-221/222 or tumor-suppressor genes such as miR-101/miR-409-3p, miR-200b/miR-200c, and miR-34a/miR-34c; its anti-colorectal cancer effect is essentially through the indirect regulation of target genes or signaling pathways. Treated by curcumin, Lnc NBR2, Lnc KCNQ1OT1, Lnc PANDAR, and Lnc CCAT1 could prove to be potentially effective target molecules in the treatment progress of colorectal cancer. Whether a large number of differentially expressed circRNAs, such as circ-3823, circ-IL4R, and circ-CUL2 in colorectal cancer, could become effective targets for curcumin in the treatment of colorectal cancer remains to be further clarified (Figure 2-ⅴ). In summary, these findings could provide favorable evidence for exploring the role of curcumin in the treatment of colorectal cancer via non-coding RNAs, which may provide new directions for the treatment and prognosis of colorectal cancer patients. Non-coding RNAs can be potential therapeutic targets for the occurrence and development of colorectal cancer, and curcumin-targeted non-coding RNAs have good biomarker and reference significance for the treatment of colorectal cancer.
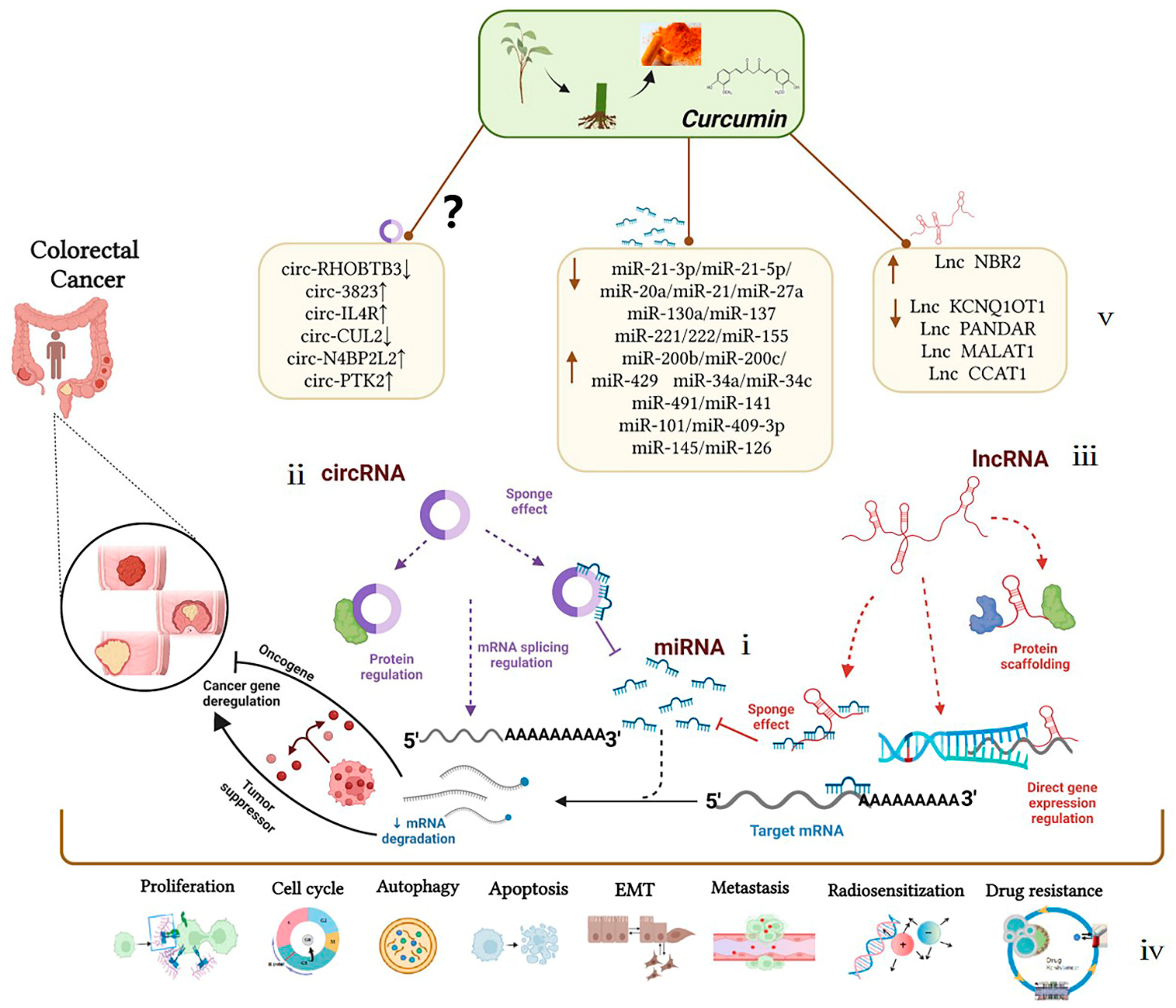
Figure 2. Mechanism of curcumin-targeted regulation of non-coding RNAs against colorectal cancer. Dotted lines represent different regulation methods; boxes represent different ncRNAs regulated by curcumin.
However, the efficacy, reliability, and sensitivity of ncRNAs as biomarkers and therapeutic targets for colorectal cancer need further basic research and clinical application. Although different types of ncRNAs have been identified to be involved in the curcumin treatment of colorectal cancer, the existing research has the following issues: (I) The regulation relationship between ncRNAs and curcumin can be found via gene knockdown and overexpression; however, whether this kind of relationship, which exists in a single cell, can be realized in the complex system of the human body still needs to be verified via further clinical trials. (II) Whether curcumin can inhibit the occurrence and development of colorectal cancer through the targeted and precise regulation of the copy number, subcellular localization, and protein binding ability of non-coding RNAs is worthy of further exploration. Additionally, the low stability, low oral bioavailability, and dose-dependent pharmacological effects of curcumin limit its clinical application in cancer therapy and industrialization. Nevertheless, curcumin is a potential candidate compound for anti-tumor drugs due to its clear biological activity and relatively simple molecular structure. The development of a new and more efficient drug delivery system of curcumin will guide its significance in the research on targeting ncRNAs and provide a new prospect for human cancer treatment [64][65]. Therefore, for better detection and effective cancer treatment, molecular diagnostic methods combined with drug treatments such as curcumin need to be researched in-depth to enrich their significance and contribute to their clinical application.
Undeniably, more differentially expressed miRNAs, lncRNAs, and circRNAs associated with colorectal cancer need to be further explored. The researchers look forward to more studies on curcumin regulating ncRNAs in colorectal cancer.
References
- Freddie Bray; Jacques Ferlay Me; Isabelle Soerjomataram; Rebecca L. Siegel; Lindsey A. Torre; Ahmedin Jemal; Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians 2018, 68, 394-424, 10.3322/caac.21492.
- Wei Cao; Hong-Da Chen; Yi-Wen Yu; Ni Li; Wan-Qing Chen; Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chinese Medical Journal 2021, 134, 783-791, 10.1097/cm9.0000000000001474.
- Hyuna Sung; Jacques Ferlay; Rebecca L. Siegel; Mathieu Laversanne; Isabelle Soerjomataram; Ahmedin Jemal; Freddie Bray; Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians 2021, 71, 209-249, 10.3322/caac.21660.
- Melina Arnold; Mónica S Sierra; Mathieu Laversanne; Isabelle Soerjomataram; Ahmedin Jemal; Freddie Bray; Global patterns and trends in colorectal cancer incidence and mortality. Gut 2016, 66, 683-691, 10.1136/gutjnl-2015-310912.
- Meng-Jia Gu; Qiu-Chi Huang; Cheng-Zhen Bao; Ying-Jun Li; Xiao-Qin Li; Ding Ye; Zhen-Hua Ye; Kun Chen; Jian-Bing Wang; Attributable causes of colorectal cancer in China. BMC Cancer 2018, 18, 1-9, 10.1186/s12885-017-3968-z.
- Changfa Xia; Xuesi Dong; He Li; Maomao Cao; Dianqin Sun; Siyi He; Fan Yang; Xinxin Yan; Shaoli Zhang; Ni Li; et al.Wanqing Chen Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chinese Medical Journal 2022, 135, 584-590, 10.1097/cm9.0000000000002108.
- Jihyoun Jeon; Mengmeng Du; Robert E. Schoen; Michael Hoffmeister; Polly A. Newcomb; Sonja I. Berndt; Bette Caan; Peter T. Campbell; Andrew T. Chan; Jenny Chang-Claude; et al.Graham G. GilesJian GongTabitha A. HarrisonJeroen R. HuygheEric J. JacobsLi LiYi LinLoïc Le MarchandJohn D. PotterConghui QuStephanie A. BienNiha ZubairRobert J. MacinnisDaniel BuchananJohn L. HopperYin CaoReiko NishiharaGad RennertMartha L. SlatteryDuncan C. ThomasMichael O. WoodsRoss L. PrenticeStephen B. GruberYingye ZhengHermann BrennerRichard HayesEmily WhiteUlrike PetersLi Hsu Determining Risk of Colorectal Cancer and Starting Age of Screening Based on Lifestyle, Environmental, and Genetic Factors. Gastroenterology 2018, 154, 2152-2164.e19, 10.1053/j.gastro.2018.02.021.
- Xuejiao Hu; Lixin Li; Mengqiao Shang; Juan Zhou; Xingbo Song; Xiaojun Lu; Jun Wang; Binwu Ying; Lanlan Wang; Association between microRNA genetic variants and susceptibility to colorectal cancer in Chinese population. Tumor Biology 2013, 35, 2151-2156, 10.1007/s13277-013-1285-y.
- Adewale Oluwaseun Fadaka; Olalekan Olanrewaju Bakare; Ashley Pretorius; Ashwil Klein; Genomic profiling of microRNA target genes in colorectal cancer. Tumor Biology 2020, 42, 1010428320933512, 10.1177/1010428320933512.
- Yufei Yang; Xuebing Yan; Xinxiang Li; Yanlei Ma; Ajay Goel; Long non-coding RNAs in colorectal cancer: Novel oncogenic mechanisms and promising clinical applications. Cancer Letters 2021, 504, 67-80, 10.1016/j.canlet.2021.01.009.
- Sepideh Mirzaei; Ali Zarrabi; Farid Hashemi; Amirhossein Zabolian; Hossein Saleki; Adnan Ranjbar; Seyed Hesam Seyed Saleh; Morteza Bagherian; Seyed Omid Sharifzadeh; Kiavash Hushmandi; et al.Alena LiskovaPeter KubatkaPooyan MakvandiVinay TergaonkarAlan Prem KumarMilad AshrafizadehGautam Sethi Regulation of Nuclear Factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis?. Cancer Letters 2021, 509, 63-80, 10.1016/j.canlet.2021.03.025.
- Yoshinaga Okugawa; William M. Grady; Ajay Goel; Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204-1225.e12, 10.1053/j.gastro.2015.07.011.
- Alexandru A. Sabo; Maria Dudau; George L. Constantin; Tudor C. Pop; Christoph-M. Geilfus; Alessio Naccarati; Mihnea P. Dragomir; Two Worlds Colliding: The Interplay Between Natural Compounds and Non-Coding Transcripts in Cancer Therapy. Frontiers in Pharmacology 2021, 12, 652074, 10.3389/fphar.2021.652074.
- Youhuang Bai; Xiaozhuan Dai; Andrew P. Harrison; Ming Chen; RNA regulatory networks in animals and plants: a long noncoding RNA perspective. Briefings in Functional Genomics 2014, 14, 91-101, 10.1093/bfgp/elu017.
- Vishnu Priya Veeraraghavan; Ullas Mony; Kaviyarasi Renu; Krishna Mohan Surapaneni; Rebai Ben Ammar; Abdullah M. AlZahrani; Emad A. Ahmed; Peramaiyan Rajendran; Effects of polyphenols on ncRNAs in cancer—An update. Clinical and Experimental Pharmacology and Physiology 2022, 49, 613-623, 10.1111/1440-1681.13641.
- Qi Yan; Fan Wu; Zhuanzhuan Yan; Jie Li; Tiantian Ma; Yufei Zhang; Yufeng Zhao; Yanrong Wang; Jiyu Zhang; Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery. BMC Plant Biology 2019, 19, 1-19, 10.1186/s12870-018-1626-5.
- Anam Javaid; Duaa Zahra; Fatima Rashid; Mutaib Mashraqi; Ahmad Alzamami; Mohsin Khurshid; Usman Ali Ashfaq; Regulation of micro-RNA, epigenetic factor by natural products for the treatment of cancers: Mechanistic insight and translational association. Saudi Journal of Biological Sciences 2022, 29, 103255, 10.1016/j.sjbs.2022.03.005.
- Kasi Pandima Devi; Tamilselvam Rajavel; Maria Daglia; Seyed Fazel Nabavi; Anupam Bishayee; Targeting miRNAs by polyphenols: Novel therapeutic strategy for cancer. Seminars in Cancer Biology 2017, 46, 146-157, 10.1016/j.semcancer.2017.02.001.
- Beilei Zhang; Ling Tian; Jinrong Xie; Guo Chen; Fu Wang; Targeting miRNAs by natural products: A new way for cancer therapy. Biomedicine & Pharmacotherapy 2020, 130, 110546, 10.1016/j.biopha.2020.110546.
- Nicola Micale; Maria Molonia; Andrea Citarella; Francesco Cimino; Antonina Saija; Mariateresa Cristani; Antonio Speciale; Natural Product-Based Hybrids as Potential Candidates for the Treatment of Cancer: Focus on Curcumin and Resveratrol. Molecules 2021, 26, 4665, 10.3390/molecules26154665.
- Man Wang; Shuai Jiang; Li Zhou; Fei Yu; Han Ding; Peifeng Li; Meng Zhou; Kun Wang; Potential Mechanisms of Action of Curcumin for Cancer Prevention: Focus on Cellular Signaling Pathways and miRNAs. International Journal of Biological Sciences 2019, 15, 1200-1214, 10.7150/ijbs.33710.
- Ammad Ahmad Farooqi; Sumbul Khalid; Aamir Ahmad; Regulation of Cell Signaling Pathways and miRNAs by Resveratrol in Different Cancers. International Journal of Molecular Sciences 2018, 19, 652, 10.3390/ijms19030652.
- Man Wang; Shuai Jiang; Fei Yu; Li Zhou; Kun Wang; Noncoding RNAs as Molecular Targets of Resveratrol Underlying Its Anticancer Effects. Journal of Agricultural and Food Chemistry 2019, 67, 4709-4719, 10.1021/acs.jafc.9b01667.
- Yu Xuan; Huiliang Yang; Linjie Zhao; Wayne Bond Lau; Bonnie Lau; Ning Ren; Yuehong Hu; Tao Yi; Xia Zhao; Shengtao Zhou; et al.Yuquan Wei MicroRNAs in colorectal cancer: Small molecules with big functions. Cancer Letters 2014, 360, 89-105, 10.1016/j.canlet.2014.11.051.
- Fangfang Yang; Guoyun Xuan; Yixin Chen; Lichao Cao; Min Zhao; Chen Wang; Erfei Chen; MicroRNAs Are Key Molecules Involved in the Gene Regulation Network of Colorectal Cancer. Frontiers in Cell and Developmental Biology 2022, 10, 828128, 10.3389/fcell.2022.828128.
- Zhen Yang; Liangcai Wu; Anqiang Wang; Wei Tang; Yi Zhao; Haitao Zhao; Andrew E. Teschendorff; dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Research 2016, 45, D812-D818, 10.1093/nar/gkw1079.
- Amir Abbas Momtazi; Fahimeh Shahabipour; Sepideh Khatibi; Thomas P. Johnston; Matteo Pirro; Amirhossein Sahebkar; Curcumin as a MicroRNA Regulator in Cancer: A Review. null 2016, 171, 1-38, 10.1007/112_2016_3.
- Siying Zhou; Sijie Zhang; Hongyu Shen; Wei Chen; Hanzi Xu; Xiu Chen; Dawei Sun; Shanliang Zhong; Jianhua Zhao; Jinhai Tang; et al. Curcumin inhibits cancer progression through regulating expression of microRNAs. Tumor Biology 2017, 39, 1010428317691680, 10.1177/1010428317691680.
- Giridhar Mudduluru; Jonahunnatha Nesson George-William; Santoshi Muppala; Irfan Asangani; Regalla Kumarswamy; Laura D. Nelson; Heike Allgayer; Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Bioscience Reports 2011, 31, 185-197, 10.1042/bsr20100065.
- Sanchita Roy; Yingjie Yu; Subhash B. Padhye; Fazlul H. Sarkar; Adhip P.N. Majumdar; Difluorinated-Curcumin (CDF) Restores PTEN Expression in Colon Cancer Cells by Down-Regulating miR-21. PLOS ONE 2013, 8, e68543, 10.1371/journal.pone.0068543.
- Huiqiang Dou; Renhui Shen; Jianxin Tao; Longchang Huang; Haoze Shi; Hang Chen; Yixin Wang; Tong Wang; Curcumin Suppresses the Colon Cancer Proliferation by Inhibiting Wnt/β-Catenin Pathways via miR-130a. Frontiers in Pharmacology 2017, 8, 877, 10.3389/fphar.2017.00877.
- Shusuke Toden; Yoshinaga Okugawa; Constanze Buhrmann; Durgha Nattamai; Esperanza Anguiano; Nicole Baldwin; Mehdi Shakibaei; C. Richard Boland; Ajay Goel; Novel Evidence for Curcumin and Boswellic Acid–Induced Chemoprevention through Regulation of miR-34a and miR-27a in Colorectal Cancer. Cancer Prevention Research 2015, 8, 431-443, 10.1158/1940-6207.capr-14-0354.
- Bai Li; Chong Shi; Bo Li; Jing‐Ming Zhao; Lei Wang; The effects of Curcumin on HCT‐116 cells proliferation and apoptosis via the miR‐491/PEG10 pathway. Journal of Cellular Biochemistry 2018, 119, 3091-3098, 10.1002/jcb.26449.
- Shusuke Toden; Yoshinaga Okugawa; Thomas Jascur; Dominik Wodarz; Natalia L. Komarova; Constanze Buhrmann; Mehdi Shakibaei; C. Richard Boland; Ajay Goel; Curcumin mediates chemosensitization to 5-fluorouracil through miRNA-induced suppression of epithelial-to-mesenchymal transition in chemoresistant colorectal cancer. Carcinogenesis 2015, 36, 355-367, 10.1093/carcin/bgv006.
- Wei Han; Hongli Yin; Hao Ma; Yi Wang; Desong Kong; Zhimin Fan; Curcumin Regulates ERCC1 Expression and Enhances Oxaliplatin Sensitivity in Resistant Colorectal Cancer Cells through Its Effects on miR-409-3p. Evidence-Based Complementary and Alternative Medicine 2020, 2020, 1-16, 10.1155/2020/8394574.
- Shruti U Gandhy; Kyounghyun Kim; Lesley Larsen; Rhonda J Rosengren; Stephen Safe; Curcumin and synthetic analogs induce reactive oxygen species and decreases specificity protein (Sp) transcription factors by targeting microRNAs. BMC Cancer 2012, 12, 564-564, 10.1186/1471-2407-12-564.
- Sanchita Roy; Edi Levi; Adhip Pn Majumdar; Fazlul H Sarkar; Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. Journal of Hematology & Oncology 2012, 5, 58-58, 10.1186/1756-8722-5-58.
- Sonia Reimondez-Troitiño; José V. González-Aramundiz; Juan Ruiz-Bañobre; Rafael López-López; María J. Alonso; Noemi Csaba; María de la Fuente; Versatile protamine nanocapsules to restore miR-145 levels and interfere tumor growth in colorectal cancer cells. European Journal of Pharmaceutics and Biopharmaceutics 2019, 142, 449-459, 10.1016/j.ejpb.2019.07.016.
- Ran Zhao; Sujuan Du; Ying Liu; Cong Lv; Yongli Song; Xinchun Chen; Bing Zhang; Dan Li; Shan Gao; Wei Cui; et al.Maksim V. PlikusXiaohua HouKaichun WuZhanju LiuZhihua LiuYingzi CongYuan LiZhengquan Yu Mucoadhesive-to-penetrating controllable peptosomes-in-microspheres co-loaded with anti-miR-31 oligonucleotide and Curcumin for targeted colorectal cancer therapy.. Theranostics 2020, 10, 3594-3611, 10.7150/thno.40318.
- Wen-Hui Fan; Feng-Chun Wang; Zhi Jin; Lin Zhu; Jian-Xin Zhang; Curcumin Synergizes with Cisplatin to Inhibit Colon Cancer through Targeting the MicroRNA-137-Glutaminase Axis. Current Medical Science 2021, 42, 108-117, 10.1007/s11596-021-2469-0.
- Ting Feng; Yumeng Wei; Robert J Lee; Ling Zhao; Liposomal curcumin and its application in cancer. International Journal of Nanomedicine 2017, ume 12, 6027-6044, 10.2147/ijn.s132434.
- Hongtao Liu; Yuan Tian; Jiaxi Li; GuoXia Zhang; Qun Liu; Min Yang; Longtao Yue; Qiwei Cao; Guihui Zhang; Yuxia Cheng; et al.Na KongLei FangShoupeng LiQing Sun Identification and functional analysis of lncRNAs and mRNAs between tumorigenesis and metastasis in CRC. Aging 2021, 13, 25859–25885, 10.18632/aging.203775.
- Qun Guang Jiang; Tai Yuan Li; Dong Ning Liu; Hai Tao Zhang; PI3K/Akt pathway involving into apoptosis and invasion in human colon cancer cells LoVo. Molecular Biology Reports 2014, 41, 3359-3367, 10.1007/s11033-014-3198-2.
- Hua Yu; Yangyang Xie; Zhendong Zhou; Zhou Wu; Xiaoyu Dai; Binbin Xu; Curcumin Regulates the Progression of Colorectal Cancer via LncRNA NBR2/AMPK Pathway.. Technology in Cancer Research & Treatment 2019, 18, 1533033819870781, 10.1177/1533033819870781.
- Zhi-Hai Zheng; He-Yi You; Yu-Jie Feng; Zhong-Tao Zhang; LncRNA KCNQ1OT1 is a Key Factor in the Reversal Effect of Curcumin on Cisplatin Resistance in the Colorectal Cancer Cells. null 2020, 476, 2575–2585, 10.21203/rs.3.rs-30554/v1.
- Tiffany Hung; Yulei Wang; Michael F Lin; Ashley K Koegel; Yojiro Kotake; Gavin Grant; Hugo M Horlings; Nilay Shah; Christopher Umbricht; Pei Wang; et al.Yu WangBenjamin KongAnita LangerødAnne-Lise Børresen-DaleSeung K KimMarc van de VijverSaraswati SukumarMichael L WhitfieldManolis KellisYue XiongDavid J WongHoward Y Chang Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genetics 2011, 43, 621-629, 10.1038/ng.848.
- Dai Wei; Li Shi Yun; Xiao Dejun; Liu Cong; Jin-Hua He; Lin Yan; Curcumin combining with si-MALAT1 inhibits the invasion and migration of colon cancer SW480 cells. Brazilian Journal of Pharmaceutical Sciences 2019, 55, 1, 10.1590/s2175-97902019000118276.
- Fan Jia; Yunhao Li; Xiongwei Deng; Xuan Wang; Xinyue Cui; Jianqing Lu; Zian Pan; Yan Wu; Self-assembled fluorescent hybrid nanoparticles-mediated collaborative lncRNA CCAT1 silencing and curcumin delivery for synchronous colorectal cancer theranostics. Journal of Nanobiotechnology 2021, 19, 1-15, 10.1186/s12951-021-00981-7.
- Shasha Zhang; Fangyi Long; Hong Lin; Xi Wang; Gang Jiang; Ting Wang; Regulatory roles of phytochemicals on circular RNAs in cancer and other chronic diseases. Pharmacological Research 2021, 174, 105936, 10.1016/j.phrs.2021.105936.
- Xiaoqing Xu; Xinyue Zhang; Yang Zhang; Zhipeng Wang; Curcumin suppresses the malignancy of non-small cell lung cancer by modulating the circ-PRKCA/miR-384/ITGB1 pathway. Biomedicine & Pharmacotherapy 2021, 138, 111439, 10.1016/j.biopha.2021.111439.
- Ji‐An Zhao; Wenjia Nie; Liang Dong; Wencong Liu; Wei Wei; A curcumin analog GL63 inhibits the malignant behaviors of hepatocellular carcinoma by inactivating the JAK2/STAT3 signaling pathway via the circular RNA zinc finger protein 83/microRNA‐324‐5p/cyclin‐dependent kinase 16 axis. Journal of Gastroenterology and Hepatology 2021, 36, 2967-2977, 10.1111/jgh.15545.
- Sifan Sun; Hailiang Fang; Curcumin inhibits ovarian cancer progression by regulating circ-PLEKHM3/miR-320a/SMG1 axis. Journal of Ovarian Research 2021, 14, 1-13, 10.1186/s13048-021-00916-8.
- Li Xue; Yuhua Tao; Yanjuan Yuan; Wei Qu; Wei Wang; Curcumin suppresses renal carcinoma tumorigenesis by regulating circ-FNDC3B/miR-138-5p/IGF2 axis. Anti-Cancer Drugs 2021, Publish Ah, 734–744, 10.1097/cad.0000000000001063.
- Bowen Du; Joong Sup Shim; Targeting Epithelial–Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules 2016, 21, 965, 10.3390/molecules21070965.
- Daoqi Zhu; Meng Shao; Jiabin Yang; Miao Fang; Shiya Liu; Dandan Lou; Ruijiao Gao; Ying Liu; Aiwu Li; Ying Lv; et al.Zhixian MoQin Fan Curcumin Enhances Radiosensitization of Nasopharyngeal Carcinoma via Mediating Regulation of Tumor Stem-like Cells by a CircRNA Network. Journal of Cancer 2020, 11, 2360-2370, 10.7150/jca.39511.
- Jiabin Yang; Daoqi Zhu; Shiya Liu; Meng Shao; Ying Liu; Aiwu Li; Ying Lv; Mu Huang; Dandan Lou; Qin Fan; et al. Curcumin enhances radiosensitization of nasopharyngeal carcinoma by regulating circRNA network. Molecular Carcinogenesis 2019, 59, 202-214, 10.1002/mc.23143.
- Jiaxin Chen; Yizheng Wu; Xin Luo; Dongai Jin; Wei Zhou; Zhenyu Ju; Di Wang; Qing Meng; Huijuan Wang; Xiaotian Fu; et al.Jianbin XuZhangfa Song Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics 2021, 11, 7507-7526, 10.7150/thno.59546.
- Yaxin Guo; Yuying Guo; Chen Chen; Dandan Fan; Xiaoke Wu; Luyang Zhao; Bo Shao; Zhenqiang Sun; Zhenyu Ji; Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Molecular Cancer 2021, 20, 1-21, 10.1186/s12943-021-01372-0.
- Tao Jiang; Hongyu Wang; Lianyu Liu; Hu Song; Yi Zhang; Jiaqi Wang; Lei Liu; Teng Xu; Ruizhi Fan; Yixin Xu; et al.Shuai WangLinsen ShiLi ZhengRenhao WangJun Song CircIL4R activates the PI3K/AKT signaling pathway via the miR-761/TRIM29/PHLPP1 axis and promotes proliferation and metastasis in colorectal cancer. Molecular Cancer 2021, 20, 1-24, 10.1186/s12943-021-01474-9.
- Ke-Da Yang; Ying Wang; Fan Zhang; Bai-Hua Luo; De-Yun Feng; Zhi-Jun Zeng; CircN4BP2L2 promotes colorectal cancer growth and metastasis through regulation of the miR-340-5p/CXCR4 axis. Laboratory Investigation 2021, 102, 38-47, 10.1038/s41374-021-00632-3.
- Bin-Lin Yang; Guo-Qiang Liu; Ping Li; Xiao-Hui Li; Circular RNA CUL2 regulates the development of colorectal cancer by modulating apoptosis and autophagy via miR-208a-3p/PPP6C. Aging 2022, 14, 497-508, 10.18632/aging.203827.
- Zhipeng Jiang; Zehui Hou; Wei Liu; Zhuomin Yu; Zhiqiang Liang; Shuang Chen; Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. Bioengineered 2022, 13, 810-823, 10.1080/21655979.2021.2012952.
- Shunhao Zhang; Jing Sun; Minqi Gu; Guihua Wang; Xudong Wang; Circular RNA: A promising new star for the diagnosis and treatment of colorectal cancer. Cancer Medicine 2021, 10, 8725-8740, 10.1002/cam4.4398.
- Aviral Kumar; Amarnath Singam; Guruprasadh Swaminathan; Naresh Killi; Naveen Kumar Tangudu; Jedy Jose; Rathna Gundloori Vn; Lekha Dinesh Kumar; Combinatorial therapy using RNAi and curcumin nano-architectures regresses tumors in breast and colon cancer models. Nanoscale 2021, 14, 492-505, 10.1039/d1nr04411g.
- Alok Vyas; Prasad Dandawate; Subhash Padhye; Aamir Ahmad; Fazlul Sarkar; Perspectives on New Synthetic Curcumin Analogs and their Potential Anticancer Properties. Current Pharmaceutical Design 2013, 19, 2047-2069, 10.2174/1381612811319110007.




