
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Wilson Ong | -- | 2896 | 2022-09-29 03:01:41 | | | |
| 2 | Lindsay Dong | Meta information modification | 2896 | 2022-09-29 03:35:44 | | |
Video Upload Options
Spinal metastasis is the most common malignant disease of the spine, and its early diagnosis and treatment is important to prevent complications and improve quality of life. The main clinical applications of AI techniques include image processing, diagnosis, decision support, treatment assistance and prognostic outcomes. In the realm of spinal oncology, artificial intelligence technologies have achieved relatively good performance and hold immense potential to aid clinicians, including enhancing work efficiency and reducing adverse events.
1. Introduction
2. Artificial Intelligence Methods for Spinal Metastasis Imaging
2.1. Artificial Intelligence (AI)
Artificial Intelligence (AI)
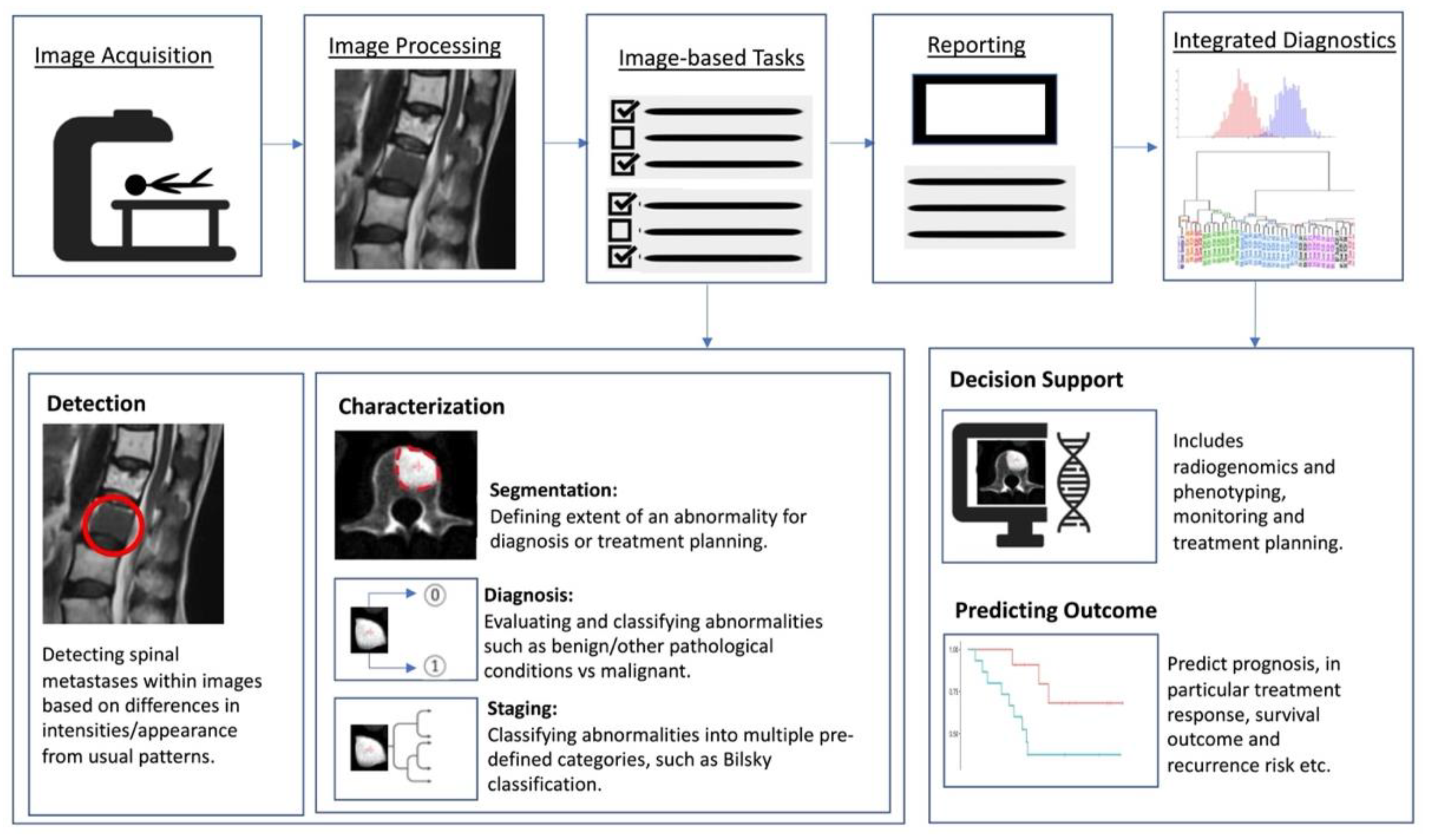
Artificial intelligence (AI) is a term referring to a machine’s computational ability to perform tasks that are comparable to those executed by humans. This is done by utilising unique inputs and then generating outputs with high added value [20]. With recent advances in medical imaging and ever increasing large amounts of digital image and report data, worldwide interest in AI for medical imaging continues to increase [21]. The rationale of using AI and computer-aided diagnostic (CAD) systems was initially thought to assist clinicians or radiologists in the detection of tumours or lesions which in turn increases efficiency, improves detection and reduces error rates [22]. As a result, efforts are ongoing to enhance the diagnostic ability of AI, and enhance its efficiency so that is can be successfully translated into clinical practice [23]. With the advent of artificial neural networks, which are a class of architectures loosely based on how the human brain works [24], several computational learning models (mainly machine learning (ML) and deep learning (DL) algorithms) have been introduced and are largely responsible for the growth of AI in radiology. In general, the clinical applications of AI (Figure 1) can be broadly characterised into three categories for oncology imaging workflow: (1) detection of abnormalities; (2) characterisation of abnormalities, which includes image processing steps such as segmentation, differentiation and classification; and (3) integrated diagnostics, which include decision support for treatment decision and planning, treatment response and prognosis prediction.
Machine Learning (ML)
Deep Learning (DL)
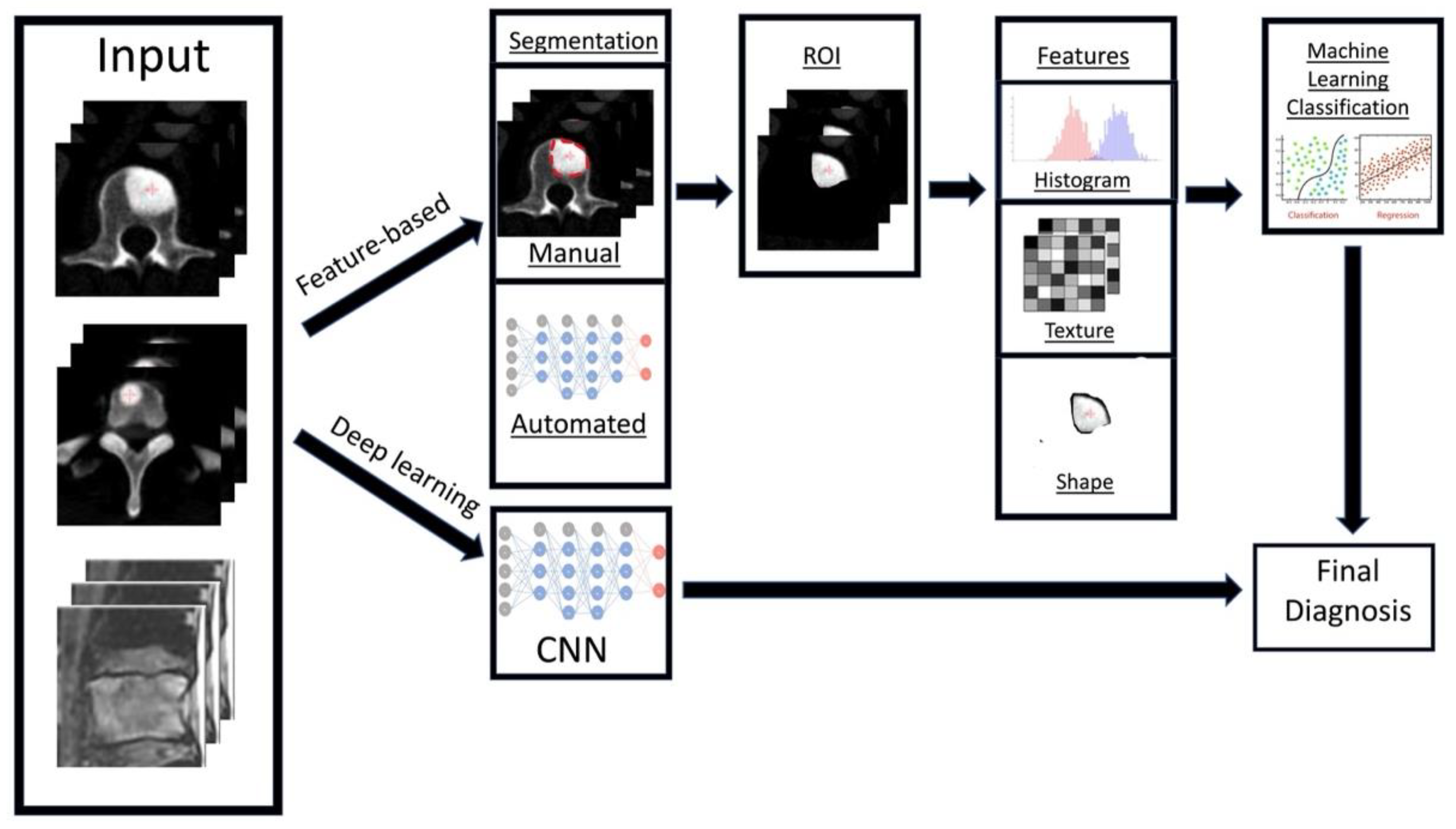
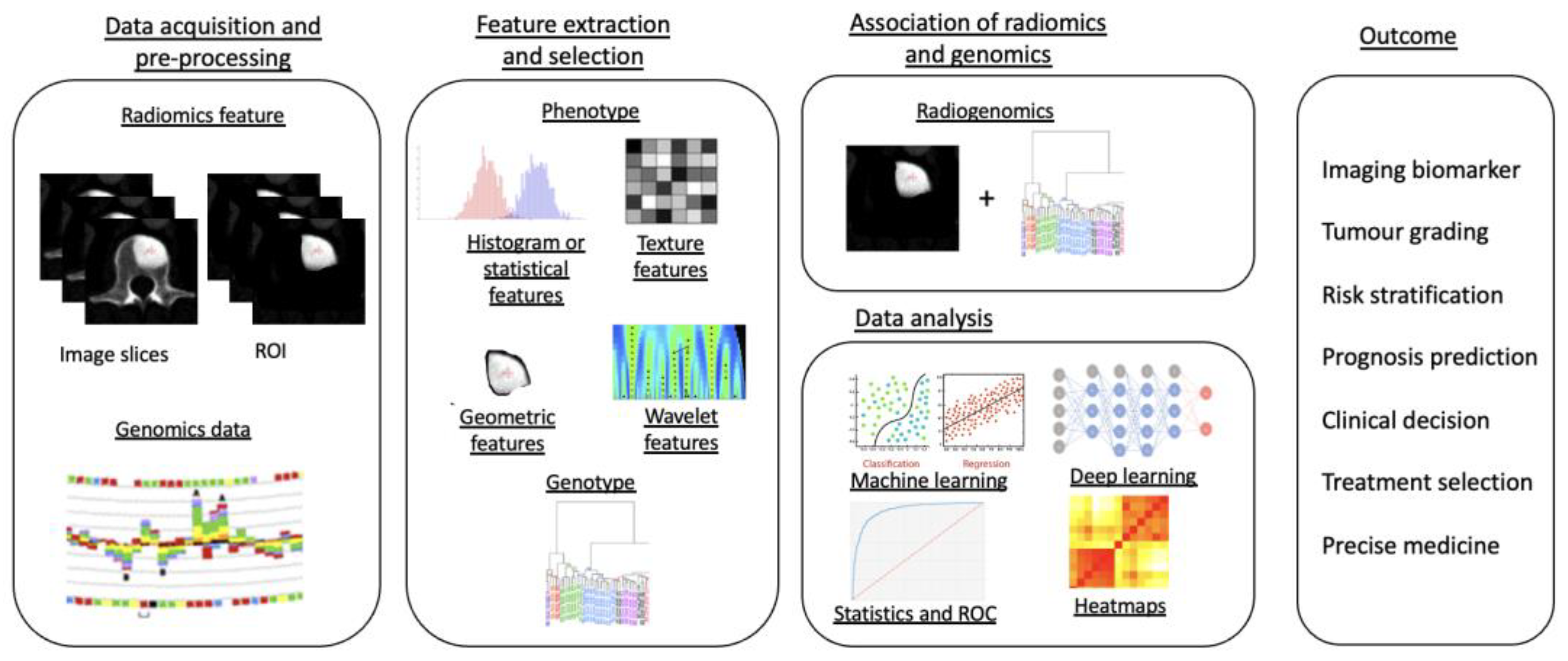
Radiomics
2.2. Artificial Intelligence Methods for Spinal Metastasis Imaging
Detection of Spinal Metastases
Differentiating Spinal Metastases from Other Pathological Conditions
Pre-Treatment Evaluation
Prediction of prognosis is a paradigm in oncological treatment. In patients with vertebral metastases, the ability to predict treatment response may help clinicians provide the most appropriate treatment with the best clinical outcome for the patient, avoid delayed transition to another treatment and prevent exposing patients to unnecessary treatment-related side effects. Shi YJ et al. [76] studied the value of MRI-based radiomics in predicting the treatment response of chemotherapy in a small group of breast cancer patients with vertebral metastases. Their radiomics model was effective in predicting progressive vs non-progressive disease with an area under the curve (AUC) of up to 0.91. This method could be extrapolated in future studies to predict the treatment response of spinal metastases and other primary tumours.
Applications of deep learning models goes beyond tumour detection and differentiation, and they have the ability to automatically generate meaningful parameters from MRI and other modalities. Hallinan et al. [77] developed a deep learning model for automated classification of metastatic epidural disease and/or spinal cord compression on MRI using the Bilsky classification. The model showed almost perfect agreement when compared to specialist readers on internal and external datasets with kappas of 0.92–0.98, p < 0.001 and 0.94–0.95, p < 0.001, respectively, for dichotomous Bilsky classification (low versus high grade). Accurate, reproducible classification of metastatic epidural spinal cord compression will enable clinicians to decide on initial radiotherapy versus surgical intervention [78].
Segmentation refers to delineation or volume extraction of a lesion or organ based on image analysis. In clinical practice, manual or semi-manual segmentation techniques are being applied to provide further value to CT and MRI studies. However, these techniques are subjective, operator-dependent and very time-consuming which limits their adoption. Automatic segmentation of spinal metastases using deep learning models has been shown to be as accurate as expert annotations in both MRI and CT [71]. Hille G et. al. [79] showed that automated vertebral metastasis segmentation on MRI using deep convolutional neural networks (U-net like architecture) were almost as accurate as expert annotation. Their automated segmentation solution achieved a Dice–Sørensen coefficient (DSC) of up to 0.78 and mean sensitivity rates up to 78.9% on par with inter-reader variability DSC of 0.79. Potentially, these models will not only reduce the need for time-consuming manual segmentation of spinal metastases, but also support stereotactic body radiotherapy planning, and improve the performance [80][81] and treatment outcome of minimally invasive interventions for spinal metastasis such as radiofrequency ablation [82]. In respect to radiotherapy, precise automated tumour contours will improve treatment planning, reduce segmentation times and reduce the radiation dose to the surrounding organs at risk, including the spinal cord. In recent years, various image segmentation techniques have been proposed, resulting in more accurate and efficient image segmentation for clinical diagnosis and treatment [83][84][85][86].
Post-Treatment Evaluation
Zhong et al. [89] created an MRI-based radiomics nomogram that was shown to be clinically useful in discriminating between cervical spine osteoradionecrosis and metastases, with an AUC of 0.73 on the training set and 0.72 in the validation set.
Pseudo-progression is a post-treatment phenomenon involving an increase in the target tumour volume (usually without any worsening symptoms), which then demonstrates interval stability or reduction in volume on repeat imaging. It occurs in approximately 14 to 18% of those with vertebral metastases treated with stereotactic body radiotherapy [90][91]. The differentiation of pseudo-progression from true progression is challenging on imaging even with many studies suggesting some differentiating factors [91][92], such as location of involvement, e.g., purely vertebral body involvement with pseudo compared to involvement of the epidural space with true progression. Artificial intelligence has already shown utility in aiding the differentiation of pseudo from true progression in brain imaging.
References
- Mundy, G.R. Metastasis to bone: Causes, consequences and therapeutic opportunities. Nat. Rev. Cancer 2002, 2, 584–593.
- Witham, T.F.; Khavkin, Y.A.; Gallia, G.L.; Wolinsky, J.P.; Gokaslan, Z.L. Surgery insight: Current management of epidural spinal cord compression from metastatic spine disease. Nat. Clin. Pract. Neurol. 2006, 2, 87–94, quiz 116.
- Klimo, P., Jr.; Schmidt, M.H. Surgical management of spinal metastases. Oncologist 2004, 9, 188–196.
- Coleman, R.E. Metastatic bone disease: Clinical features, pathophysiology and treatment strategies. Cancer Treat Rev. 2001, 27, 165–176.
- Cuccurullo, V.; Cascini, G.L.; Tamburrini, O.; Rotondo, A.; Mansi, L. Bone metastases radiopharmaceuticals: An overview. Curr. Radiopharm. 2013, 6, 41–47.
- Cecchini, M.G.; Wetterwald, A.; Pluijm, G.v.d.; Thalmann, G.N. Molecular and Biological Mechanisms of Bone Metastasis. EAU Update Ser. 2005, 3, 214–226.
- Yu, H.H.; Tsai, Y.Y.; Hoffe, S.E. Overview of diagnosis and management of metastatic disease to bone. Cancer Control 2012, 19, 84–91.
- O’Sullivan, G.J.; Carty, F.L.; Cronin, C.G. Imaging of bone metastasis: An update. World J. Radiol. 2015, 7, 202–211.
- Liu, T.; Wang, S.; Liu, H.; Meng, B.; Zhou, F.; He, F.; Shi, X.; Yang, H. Detection of vertebral metastases: A meta-analysis comparing MRI, CT, PET, BS and BS with SPECT. J. Cancer Res. Clin. Oncol. 2017, 143, 457–465.
- Wallace, A.N.; Greenwood, T.J.; Jennings, J.W. Use of Imaging in the Management of Metastatic Spine Disease With Percutaneous Ablation and Vertebral Augmentation. AJR Am. J. Roentgenol. 2015, 205, 434–441.
- Moynagh, M.R.; Colleran, G.C.; Tavernaraki, K.; Eustace, S.J.; Kavanagh, E.C. Whole-body magnetic resonance imaging: Assessment of skeletal metastases. Semin. Musculoskelet. Radiol. 2010, 14, 22–36.
- Schiff, D.; O’Neill, B.P.; Wang, C.H.; O’Fallon, J.R. Neuroimaging and treatment implications of patients with multiple epidural spinal metastases. Cancer 1998, 83, 1593–1601.
- Talbot, J.N.; Paycha, F.; Balogova, S. Diagnosis of bone metastasis: Recent comparative studies of imaging modalities. Q. J. Nucl. Med. Mol. Imaging 2011, 55, 374–410.
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J.V.; Waddell, N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021, 13, 152.
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics 2019, 9, 1303–1322.
- Parekh, V.S.; Jacobs, M.A. Deep learning and radiomics in precision medicine. Expert Rev. Precis Med. Drug Dev. 2019, 4, 59–72.
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118.
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131 e1129.
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350.
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358.
- Thrall, J.H.; Li, X.; Li, Q.; Cruz, C.; Do, S.; Dreyer, K.; Brink, J. Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J. Am. Coll Radiol. 2018, 15, 504–508.
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510.
- Nagoev, Z.V.; Sundukov, Z.A.; Pshenokova, I.A.; Denisenko, V.A. Architecture of CAD for distributed artificial intelligence based on self-organizing neuro-cognitive architectures. News Kabard.–Balkar Sci. Cent. RAS 2020, 2, 40–47.
- Kriegeskorte, N. Deep Neural Networks: A New Framework for Modeling Biological Vision and Brain Information Processing. Annu. Rev. Vis. Sci. 2015, 1, 417–446.
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515.
- Zhu, X.; Goldberg, A.B. Introduction to Semi-Supervised Learning. Synth. Lect. Artif. Intell. Mach. Learn. 2009, 3, 1–130.
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64.
- Cao, B.; Araujo, A.; Sim, J. Unifying Deep Local and Global Features for Image Search. In Proceedings of the Computer Vision–ECCV 2020, Glasgow, UK, 23–28 August 2020; pp. 726–743.
- Bengio, Y. Learning Deep Architectures for AI. Found. Trends Mach. Learn. 2009, 2, 1–127.
- Montagnon, E.; Cerny, M.; Cadrin-Chênevert, A.; Hamilton, V.; Derennes, T.; Ilinca, A.; Vandenbroucke-Menu, F.; Turcotte, S.; Kadoury, S.; Tang, A. Deep learning workflow in radiology: A primer. Insights Imaging 2020, 11, 22.
- Li, H.; Galperin-Aizenberg, M.; Pryma, D.; Simone, C.B., 2nd; Fan, Y. Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy. Radiother. Oncol. 2018, 129, 218–226.
- Alaverdyan, Z.; Jung, J.; Bouet, R.; Lartizien, C. Regularized siamese neural network for unsupervised outlier detection on brain multiparametric magnetic resonance imaging: Application to epilepsy lesion screening. Med. Image Anal. 2020, 60, 101618.
- Tlusty, T.; Amit, G.; Ben-Ari, R. Unsupervised clustering of mammograms for outlier detection and breast density estimation. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3808–3813.
- Zaharchuk, G.; Gong, E.; Wintermark, M.; Rubin, D.; Langlotz, C.P. Deep Learning in Neuroradiology. AJNR Am. J. Neuroradiol. 2018, 39, 1776–1784.
- Kaka, H.; Zhang, E.; Khan, N. Artificial intelligence and deep learning in neuroradiology: Exploring the new frontier. Can. Assoc. Radiol. J. 2021, 72, 35–44.
- Cheng, P.M.; Montagnon, E.; Yamashita, R.; Pan, I.; Cadrin-Chênevert, A.; Romero, F.P.; Chartrand, G.; Kadoury, S.; Tang, A. Deep Learning: An Update for Radiologists. RadioGraphics 2021, 41, 1427–1445.
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging 2020, 11, 91.
- Faiella, E.; Santucci, D.; Calabrese, A.; Russo, F.; Vadala, G.; Zobel, B.B.; Soda, P.; Iannello, G.; de Felice, C.; Denaro, V. Artificial Intelligence in Bone Metastases: An MRI and CT Imaging Review. Int. J. Environ. Res. Public Health 2022, 19, 1880.
- Mannil, M.; von Spiczak, J.; Manka, R.; Alkadhi, H. Texture Analysis and Machine Learning for Detecting Myocardial Infarction in Noncontrast Low-Dose Computed Tomography: Unveiling the Invisible. Investig. Radiol. 2018, 53, 338–343.
- Aerts, H.J. The Potential of Radiomic-Based Phenotyping in Precision Medicine: A Review. JAMA Oncol. 2016, 2, 1636–1642.
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446.
- Valladares, A.; Beyer, T.; Rausch, I. Physical imaging phantoms for simulation of tumor heterogeneity in PET, CT, and MRI: An overview of existing designs. Med. Phys. 2020, 47, 2023–2037.
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32.
- Zhang, Y.; Oikonomou, A.; Wong, A.; Haider, M.A.; Khalvati, F. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, 46349.
- Jia, T.-Y.; Xiong, J.-F.; Li, X.-Y.; Yu, W.; Xu, Z.-Y.; Cai, X.-W.; Ma, J.-C.; Ren, Y.-C.; Larsson, R.; Zhang, J.; et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur. Radiol. 2019, 29, 4742–4750.
- Rogers, W.; Thulasi Seetha, S.; Refaee, T.A.G.; Lieverse, R.I.Y.; Granzier, R.W.Y.; Ibrahim, A.; Keek, S.A.; Sanduleanu, S.; Primakov, S.P.; Beuque, M.P.L.; et al. Radiomics: From qualitative to quantitative imaging. Br. J. Radiol. 2020, 93, 20190948.
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36.
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762.
- Zhou, M.; Scott, J.; Chaudhury, B.; Hall, L.; Goldgof, D.; Yeom, K.W.; Iv, M.; Ou, Y.; Kalpathy-Cramer, J.; Napel, S.; et al. Radiomics in Brain Tumor: Image Assessment, Quantitative Feature Descriptors, and Machine-Learning Approaches. AJNR Am. J. Neuroradiol. 2018, 39, 208–216.
- Chen, C.; Zheng, A.; Ou, X.; Wang, J.; Ma, X. Comparison of Radiomics-Based Machine-Learning Classifiers in Diagnosis of Glioblastoma From Primary Central Nervous System Lymphoma. Front. Oncol. 2020, 10, 1151.
- Cha, Y.J.; Jang, W.I.; Kim, M.S.; Yoo, H.J.; Paik, E.K.; Jeong, H.K.; Youn, S.M. Prediction of Response to Stereotactic Radiosurgery for Brain Metastases Using Convolutional Neural Networks. Anticancer Res. 2018, 38, 5437–5445.
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48.
- Ciresan, D.C.; Meier, U.; Masci, J.; Gambardella, L.M.; Schmidhuber, J. Flexible, high performance convolutional neural networks for image classification. In Proceedings of the Twenty-Second International Joint Conference on Artificial Intelligence, Barcelona, Spain, 16–22 July 2011.
- Papadimitroulas, P.; Brocki, L.; Christopher Chung, N.; Marchadour, W.; Vermet, F.; Gaubert, L.; Eleftheriadis, V.; Plachouris, D.; Visvikis, D.; Kagadis, G.C.; et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys. Med. 2021, 83, 108–121.
- Ueda, D.; Shimazaki, A.; Miki, Y. Technical and clinical overview of deep learning in radiology. Jpn. J. Radiol. 2019, 37, 15–33.
- Haider, S.P.; Burtness, B.; Yarbrough, W.G.; Payabvash, S. Applications of radiomics in precision diagnosis, prognostication and treatment planning of head and neck squamous cell carcinomas. Cancers Head Neck 2020, 5, 6.
- Curtin, M.; Piggott, R.P.; Murphy, E.P.; Munigangaiah, S.; Baker, J.F.; McCabe, J.P.; Devitt, A. Spinal Metastatic Disease: A Review of the Role of the Multidisciplinary Team. Orthop. Surg. 2017, 9, 145–151.
- Tomita, K.; Kawahara, N.; Kobayashi, T.; Yoshida, A.; Murakami, H.; Akamaru, T. Surgical strategy for spinal metastases. Spine (Phila Pa 1976) 2001, 26, 298–306.
- Clemons, M.; Gelmon, K.A.; Pritchard, K.I.; Paterson, A.H. Bone-targeted agents and skeletal-related events in breast cancer patients with bone metastases: The state of the art. Curr. Oncol. 2012, 19, 259–268.
- Hamaoka, T.; Madewell, J.E.; Podoloff, D.A.; Hortobagyi, G.N.; Ueno, N.T. Bone imaging in metastatic breast cancer. J. Clin. Oncol. 2004, 22, 2942–2953.
- Bilsky, M.H.; Lis, E.; Raizer, J.; Lee, H.; Boland, P. The diagnosis and treatment of metastatic spinal tumor. Oncologist 1999, 4, 459–469.
- Burns, J.E.; Yao, J.; Wiese, T.S.; Munoz, H.E.; Jones, E.C.; Summers, R.M. Automated detection of sclerotic metastases in the thoracolumbar spine at CT. Radiology 2013, 268, 69–78.
- Hammon, M.; Dankerl, P.; Tsymbal, A.; Wels, M.; Kelm, M.; May, M.; Suehling, M.; Uder, M.; Cavallaro, A. Automatic detection of lytic and blastic thoracolumbar spine metastases on computed tomography. Eur. Radiol. 2013, 23, 1862–1870.
- O’Connor, S.D.; Yao, J.; Summers, R.M. Lytic metastases in thoracolumbar spine: Computer-aided detection at CT–preliminary study. Radiology 2007, 242, 811–816.
- Wiese, T.; Yao, J.; Burns, J.E.; Summers, R.M. Detection of sclerotic bone metastases in the spine using watershed algorithm and graph cut. In Proceedings of the Medical Imaging 2012: Computer-Aided Diagnosis, San Diego, CA, USA, 4–9 February 2012.
- Aneja, S.; Chang, E.; Omuro, A. Applications of artificial intelligence in neuro-oncology. Curr. Opin. Neurol 2019, 32, 850–856.
- Duong, M.T.; Rauschecker, A.M.; Mohan, S. Diverse Applications of Artificial Intelligence in Neuroradiology. Neuroimaging Clin. N. Am. 2020, 30, 505–516.
- Muthukrishnan, N.; Maleki, F.; Ovens, K.; Reinhold, C.; Forghani, B.; Forghani, R. Brief History of Artificial Intelligence. Neuroimaging Clin. N. Am. 2020, 30, 393–399.
- Wang, J.; Fang, Z.; Lang, N.; Yuan, H.; Su, M.Y.; Baldi, P. A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Comput. Biol. Med. 2017, 84, 137–146.
- Fan, X.; Zhang, X.; Zhang, Z.; Jiang, Y. Deep Learning-Based Identification of Spinal Metastasis in Lung Cancer Using Spectral CT Images. Sci. Program. 2021, 2021, 2779390.
- Chang, C.Y.; Buckless, C.; Yeh, K.J.; Torriani, M. Automated detection and segmentation of sclerotic spinal lesions on body CTs using a deep convolutional neural network. Skelet. Radiol. 2022, 51, 391–399.
- Roth, H.Y.J.; Lu, L.; Stieger, J.; Burns, J.; Summers, R.M. Detection of Sclerotic Spine Metastases via Random Aggregation of Deep Convolutional Neural Network Classifications. Lect. Notes Comput. Vis. Biomech. 2014, 20, 3–12.
- Filograna, L.; Lenkowicz, J.; Cellini, F.; Dinapoli, N.; Manfrida, S.; Magarelli, N.; Leone, A.; Colosimo, C.; Valentini, V. Identification of the most significant magnetic resonance imaging (MRI) radiomic features in oncological patients with vertebral bone marrow metastatic disease: A feasibility study. Radiol. Med. 2019, 124, 50–57.
- Liu, J.; Guo, W.; Zeng, P.; Geng, Y.; Liu, Y.; Ouyang, H.; Lang, N.; Yuan, H. Vertebral MRI-based radiomics model to differentiate multiple myeloma from metastases: Influence of features number on logistic regression model performance. Eur. Radiol. 2022, 32, 572–581.
- Xiong, X.; Wang, J.; Hu, S.; Dai, Y.; Zhang, Y.; Hu, C. Differentiating Between Multiple Myeloma and Metastasis Subtypes of Lumbar Vertebra Lesions Using Machine Learning-Based Radiomics. Front. Oncol. 2021, 11, 601699.
- Shi, Y.J.; Zhu, H.T.; Li, X.T.; Zhang, X.Y.; Wei, Y.Y.; Yan, S.; Sun, Y.S. Radiomics analysis based on multiple parameters MR imaging in the spine: Predicting treatment response of osteolytic bone metastases to chemotherapy in breast cancer patients. Magn. Reson. Imaging 2022, 92, 10–18.
- Hallinan, J.T.P.D.; Zhu, L.; Zhang, W.; Lim, D.S.W.; Baskar, S.; Low, X.Z.; Yeong, K.Y.; Teo, E.C.; Kumarakulasinghe, N.B.; Yap, Q.V.; et al. Deep Learning Model for Classifying Metastatic Epidural Spinal Cord Compression on MRI. Front. Oncol. 2022, 12.
- Perry, J.; Chambers, A.; Laperriere, N. Systematic Review of the Diagnosis and Management of Malignant Extradural Spinal Cord Compression: The Cancer Care Ontario Practice Guidelines Initiative‘s Neuro-Oncology Disease Site Group. J. Clin. Oncol. 2005, 23, 2028–2037.
- Hille, G.; Steffen, J.; Dünnwald, M.; Becker, M.; Saalfeld, S.; Tönnies, K. Spinal Metastases Segmentation in MR Imaging using Deep Convolutional Neural Networks. arXiv 2020, arXiv:2001.05834.
- Boon, I.S.; Au Yong, T.P.T.; Boon, C.S. Assessing the Role of Artificial Intelligence (AI) in Clinical Oncology: Utility of Machine Learning in Radiotherapy Target Volume Delineation. Medicines 2018, 5, 131.
- Li, Q.; Xu, Y.; Chen, Z.; Liu, D.; Feng, S.T.; Law, M.; Ye, Y.; Huang, B. Tumor Segmentation in Contrast-Enhanced Magnetic Resonance Imaging for Nasopharyngeal Carcinoma: Deep Learning with Convolutional Neural Network. BioMed Res. Int. 2018, 2018, 9128527.
- Arends, S.R.S.; Savenije, M.H.F.; Eppinga, W.S.C.; van der Velden, J.M.; van den Berg, C.A.T.; Verhoeff, J.J.C. Clinical utility of convolutional neural networks for treatment planning in radiotherapy for spinal metastases. Phys. Imaging Radiat. Oncol. 2022, 21, 42–47.
- Wong, J.; Fong, A.; McVicar, N.; Smith, S.; Giambattista, J.; Wells, D.; Kolbeck, C.; Giambattista, J.; Gondara, L.; Alexander, A. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother. Oncol. 2020, 144, 152–158.
- Wang, Z.; Chang, Y.; Peng, Z.; Lv, Y.; Shi, W.; Wang, F.; Pei, X.; Xu, X.G. Evaluation of deep learning-based auto-segmentation algorithms for delineating clinical target volume and organs at risk involving data for 125 cervical cancer patients. J. Appl. Clin. Med. Phys. 2020, 21, 272–279.
- Men, K.; Dai, J.; Li, Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med. Phys. 2017, 44, 6377–6389.
- Vrtovec, T.; Močnik, D.; Strojan, P.; Pernuš, F.; Ibragimov, B. Auto-segmentation of organs at risk for head and neck radiotherapy planning: From atlas-based to deep learning methods. Med. Phys. 2020, 47, e929–e950.
- Saxena, S.; Jena, B.; Gupta, N.; Das, S.; Sarmah, D.; Bhattacharya, P.; Nath, T.; Paul, S.; Fouda, M.M.; Kalra, M.; et al. Role of Artificial Intelligence in Radiogenomics for Cancers in the Era of Precision Medicine. Cancers 2022, 14, 2860.
- Fathi Kazerooni, A.; Bagley, S.J.; Akbari, H.; Saxena, S.; Bagheri, S.; Guo, J.; Chawla, S.; Nabavizadeh, A.; Mohan, S.; Bakas, S.; et al. Applications of Radiomics and Radiogenomics in High-Grade Gliomas in the Era of Precision Medicine. Cancers 2021, 13, 5921.
- Zhong, X.; Li, L.; Jiang, H.; Yin, J.; Lu, B.; Han, W.; Li, J.; Zhang, J. Cervical spine osteoradionecrosis or bone metastasis after radiotherapy for nasopharyngeal carcinoma? The MRI-based radiomics for characterization. BMC Med. Imaging 2020, 20, 104.
- Amini, B.; Beaman, C.B.; Madewell, J.E.; Allen, P.K.; Rhines, L.D.; Tatsui, C.E.; Tannir, N.M.; Li, J.; Brown, P.D.; Ghia, A.J. Osseous Pseudoprogression in Vertebral Bodies Treated with Stereotactic Radiosurgery: A Secondary Analysis of Prospective Phase I/II Clinical Trials. AJNR Am. J. Neuroradiol. 2016, 37, 387–392.
- Bahig, H.; Simard, D.; Letourneau, L.; Wong, P.; Roberge, D.; Filion, E.; Donath, D.; Sahgal, A.; Masucci, L. A Study of Pseudoprogression After Spine Stereotactic Body Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 848–856.
- Taylor, D.R.; Weaver, J.A. Tumor pseudoprogression of spinal metastasis after radiosurgery: A novel concept and case reports. J. Neurosurg. Spine 2015, 22, 534–539.




