Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Meriem Miyassa Aci | -- | 2843 | 2022-09-26 14:09:32 | | | |
| 2 | Camila Xu | -11 word(s) | 2832 | 2022-09-27 03:02:22 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Aci, M.M.; Sidari, R.; Araniti, F.; Lupini, A. Genomic Approaches in Allelopathy. Encyclopedia. Available online: https://encyclopedia.pub/entry/27598 (accessed on 07 February 2026).
Aci MM, Sidari R, Araniti F, Lupini A. Genomic Approaches in Allelopathy. Encyclopedia. Available at: https://encyclopedia.pub/entry/27598. Accessed February 07, 2026.
Aci, Meriem Miyassa, Rossana Sidari, Fabrizio Araniti, Antonio Lupini. "Genomic Approaches in Allelopathy" Encyclopedia, https://encyclopedia.pub/entry/27598 (accessed February 07, 2026).
Aci, M.M., Sidari, R., Araniti, F., & Lupini, A. (2022, September 26). Genomic Approaches in Allelopathy. In Encyclopedia. https://encyclopedia.pub/entry/27598
Aci, Meriem Miyassa, et al. "Genomic Approaches in Allelopathy." Encyclopedia. Web. 26 September, 2022.
Copy Citation
Allelopathy is an ecological phenomenon, in which the chemicals produced by plants and microorganisms affect the growth, development, and fitness of other organisms. This discipline represents a topic of growing interest due to the sustainability discussion currently in progress. Genetic technologies have developed rapidly, and whole-genome sequencing has become an increasingly routine technique in many areas, such as medicine, biotechnology, and agriculture.
allelopathic genes
quantitative trait loci (QTL)
weed control
microorganisms
Next Generation Sequencing
Cross-kingdome RNAi
1. Introduction
Allelopathy is an ecological phenomenon, in which the chemicals produced by plants and microorganisms affect the growth, development, and fitness of other organisms [1]. This discipline represents a topic of growing interest due to the sustainability discussion currently in progress [2]. Over the years, several definitions have been adopted, in which “interaction” has been the key common word. Many definitions of allelopathy have been given throughout history [3][4][5]. More recently, the International Allelopathy Society (IAS) has further expanded the definition as follows: “any process involving secondary metabolites produced by plants, microorganisms, viruses, and fungi that influence the growth and development of agricultural and biological systems” (IAS, 1996) (Figure 1). However, although the different definitions mentioned above have tried to include all the possible physiological responses due to allelopathic interactions induced by secondary metabolites among organisms, to date the positive or negative effects of allelopathy are not well defined [5].
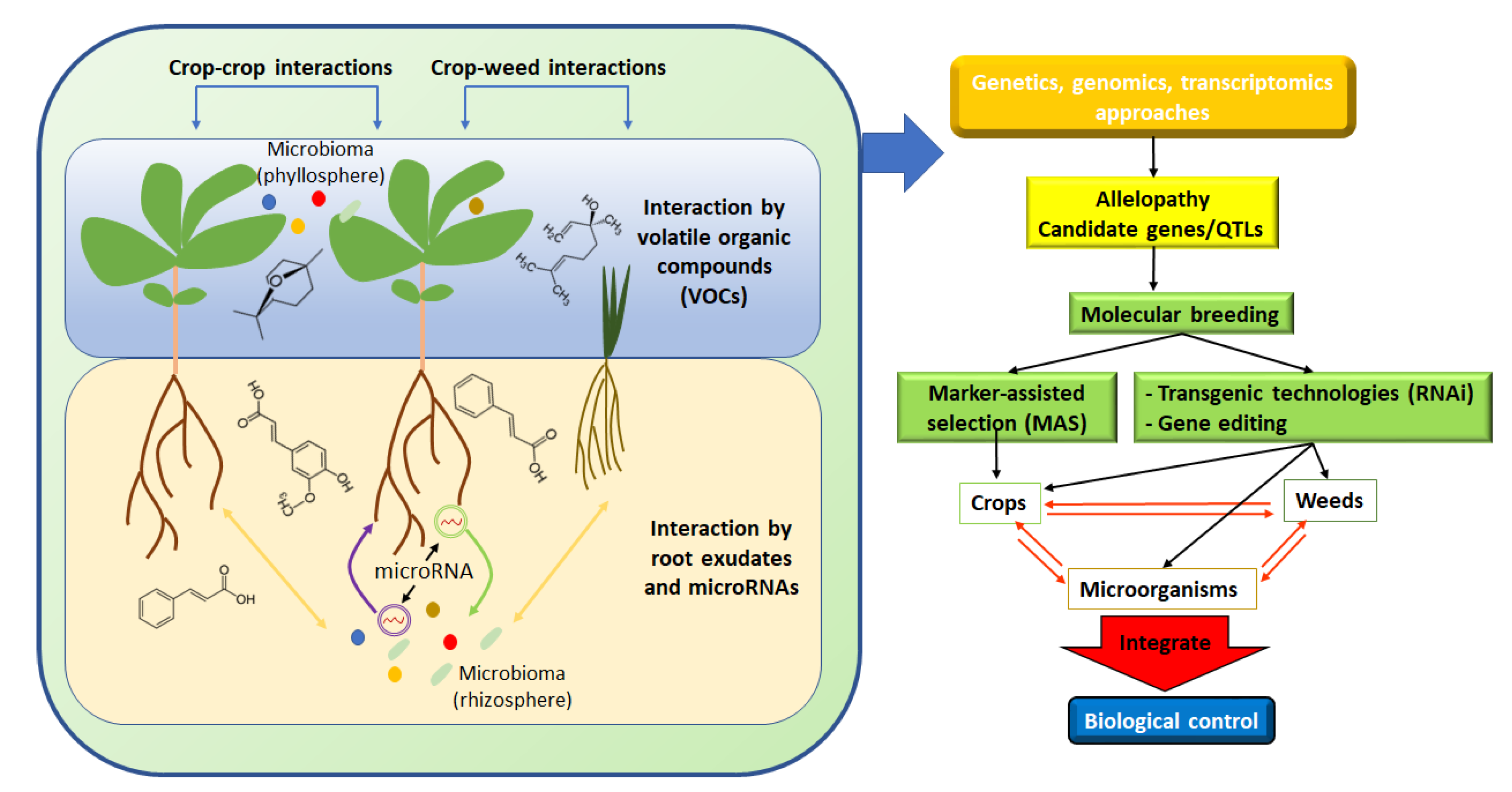
Figure 1. Schematic diagram highlighting the different allelopathic interactions. Aboveground interactions are mediated by volatile organic compounds (VOCs), whereas root exudates represent the main factor mediating allelopathic interaction in the soil. Two strategies (transgenic organisms or MAS) can be pursued to create new varieties with higher allelopathic potential. Microbiota modulates, reduces, or enhances, allelopathic interactions even through cross-kingdom microRNA exchanges.
Indeed, the study of plant responses to allelochemicals is markedly influenced by the used methods, the considered biological traits, and the evolutionary history of the organisms [5]. In addition, the allelopathic phenomenon increases with the genetic distance between the species, and this concept opens a new scenario in which kin recognition takes place among species [6] and where root exudates play a pivotal role [7]. In this context, Crepy and Casal [8] described for the first time the molecular mechanisms of recognition responses in the shoot in which phytochrome B and cryptochrome 1 genes were involved. Moreover, plants can be passive organisms, responding only to environmental fluctuations, or active, transmitting, receiving, and reacting directly with other plants and microorganisms to chemical signals, regardless of environmental variations (Figure 1) [9]. These responses, which determine a fundamental role in the acquisition of resources, are the key to how a plant community is organized and how species-specific mechanisms, such as coevolution, are modulated [10][11][12].
In field conditions, the allelopathic phenomenon can also be easily understood from the spatial distribution of species [13][14][15][16][17]. taking into account the architecture of the root system as well [18]. Moreover, for many years, allelopathy was considered an aspect of the plant competition phenomenon, but today, the distinction between the two phenomena is very clear [19][20].
Allelochemicals are secondary metabolites from different classes, such as phenolic, terpenoid, and alkaloid compounds [21]. As the main allelochemicals within plants, in terms of proportion, the phenolic compounds were extensively studied to identify their allelopathic mechanism of action in model species, such as Arabidopsis thaliana and Lactuca sativa, although no field applications have been performed [21]. Several studies reported the allelopathic effect of simple phenolic compounds on the morphophysiological processes in many crops, such as root morphology in maize plants [22], membrane permeability [23], nutrient uptake [22], cell division and elongation [24], photosynthesis and respiration [25], and hormones synthesis and balance [26].
On the other hand, more complex metabolites were also studied in allelopathic interaction such as terpenoids, including mono-, di-, triterpenoids, and sterols [21], which are involved in seed germination and oxidative damage [26], plant communication with other organisms [27], and plant defense as well [28].
Moreover, in agroecosystems, the allelopathic effects take on considerable importance in weed management [29]. Indeed, among crop pests, agricultural weeds represent the major limitation to agricultural production [30], and chemical weed control currently represents the most adopted strategy, leading to environment and human health cues [31]. A strategy to overcome these problems could be the use of allelochemicals, which possess a high potential as bioherbicide or/and herbicide bioinspired, exploiting new mechanisms of action, thereby overcoming specific resistances [12], thus representing an alternative to weed control in terms of sustainability.
2. Genomic Approaches in Allelopathy
In recent years, genetic technologies have developed rapidly, and whole-genome sequencing has become an increasingly routine technique in many areas, such as medicine, biotechnology, and agriculture. Although global expression responses of plant genomes in allelopathy using DNA microarrays were reported [32], the new technologies could detect novel transcripts, and predict the gene regulatory networks of a biological response. Indeed, to analyze gene and/or regulatory chromosome regions, a complete sequence assembly is necessary, and, at the same time, the biological processes associated with likely phenotypic traits can be determined. To date, these sequencing technologies, such as whole-genome sequencing and RNA sequencing (RNA-seq), can be performed at different biological levels: from plant tissue to single cell [33]. However, in allelopathy, the application of these approaches is markedly limited to a few experiments (Table 1), including either gene expression or RNA sequencing technologies, as well as the identification of quantitative trait loci (QTL) useful for plant breeding.
Table 1. Representative articles of the genetic and genomic approaches in allelopathy.
| Plant Material | Methods | Targets | References |
|---|---|---|---|
| Common reed | RNA-seq | Phytohormones | He et al. [34] |
| Rice/barnyard grass | Microarray | Phytohormones | Chi et al. [35] |
| Tomato | RNA-seq | Antioxidants and Hormones | Cheng et al. [36] |
| Rice/barnyard grass | RNA-seq | Shikimic acid and acetic acid pathways | Zhang et al. [37] |
| Rice/barnyard grass | RNA-seq | Diterpenoid and flavonoid biosynthesis pathway | Li et al. [38] |
| Rehmannia glutinosa | Cloning, qRT-PCR | Phenolic biosynthesis: C3H gene | Yang et al. [39] |
| Soybeans | RNA-seq | Oxidative stress and jasmonic acid signaling (PIF3) |
Horvath et al. [40] |
| Rice | SNPs genotyping | QTL regions | Chung et al. [41] |
| Rice | qRT-PCR | Biosynthesis of phenolic acids | Zhang et al. [42] |
| Wheat/ryegrass | AFLP, RFLP and SSR genotyping | QTL regions | Wu et al. [43] |
| Lettuce/rice | RFLP genotyping | QTL regions | Zeng et al. [44] |
| Lettuce/Triticum Speltoides | RAPD genotyping | Genetic diversity in allelopathic potential | Quader et al. [45] |
| Rice | RNA interference | PAL gene expression | Fang et al. [46] |
| Rice | T-DNA insertion | OsCPS4, OsKSL4 | Xu et al. [47] |
| Rice/barnyard grass | qRT-PCR | PAL, C4H, F5H, and COMT genes | Zhang et al. [48] |
| Sorghum | SSR genotyping | QTL regions | Shehzad et al. [49] |
| Rice/barnyard grass | qRT-PCR, ChIP-seq, ChIP-qPCR | MYB transcription factor | Fang et al. [50] |
| Rice | qRT-PCR, RNA-seq | Biosynthetic gene clusters | Sultana et al. [51] |
| Arabidopsis | RNA-seq | Signal transduction, nutrient transporter, detoxification genes | Zhang et al. [52] |
| Rice | RNA-seq | Chlorophyll and nitrogen metabolisms | Li et al. [53] |
| Rice | Genome sequencing | detoxification-related genes (CYP450, GST) DIMBOA gene cluster. | Guo et al. [54] |
2.1. From Metabolite to Gene in Plants
Despite the considerable number of manuscripts dealing with crop allelopathy, the main works aiming to understand the molecular networks involved in allelopathic traits and their potential involvement in breeding programs for field application were performed on cereals (Table 1). In particular, the main metabolites considered to be involved in this phenomenon belong to the classes of indoles (benzoxazinoids and their derivatives, produced by wheat, maize, rice, etc.), phenylpropanoids (i.e., cinnamic acids derivatives), and terpenoids (momilactones a and b, produced by rice).
Among the molecular approaches used to clarify some aspects related to the allelopathic phenomenon, the expression analyses of plant secondary metabolism pathway-related genes turned out to be an informative technique for the regulation of metabolite biosynthesis. The first correlation between metabolite produced and gene expression was demonstrated in cereals [46][55]. In particular, some studies focused on the biosynthesis of benzoxazinoide compounds, the cyclic hydroxamic acids 2,4-dihydroxy-1,4-benzoxazin-3-one and 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIBOA and DIMBOA, respectively), identifying five key responsible genes in maize [56]. In this regard, a mutation in the Bx1 gene in maize demonstrated a clear correlation between gene (Bx1) and metabolite (DIMBOA). Then, a deeper study of the same biosynthetic pathway led to the identification of other genes (named Bx2 through Bx5) encoding cytochrome P450-dependent monooxygenases [56]. Similarly, Song et al. [57] investigated in hydroponic experiments the weed suppressive ability of several rice accessions and banyardgrass (Echinochloa crus-galli) exposed to different nitrogen supplies, identifying two contrasting lines (accession PI312777, highly allelopathic, and accession Lemont, with low suppressive activity). After a subtractive hybridization suppression, to construct a forward SSH-cDNA library of PI312777, the researchers sequenced and annotated 35 clones, identifying genes related to allelochemicals. In particular, they reported that in the accession PI312777, the phenylalanine ammonia lyase (PAL) and cytochrome P450 genes strongly increased their transcript abundance at a low N level, suggesting that the higher ability of PI312777 to suppress banyardgrass might be connected to the stronger activation of genes involved in de novo synthesis of allelochemicals [57].
Besides gene expression and transcriptomic analysis, the involvement of PAL-related genes in rice allelopathy was further elucidated using proteomics and bioinformatics approaches. A significant correlation between inhibitory effects of allelopathic rice on weeds and a higher expression of PAL in the phenylpropanoid metabolism was demonstrated using the RNA interference (RNAi) approach to silence this gene in rice, highlighting also a quali-quantitative modulation of microorganisms in the rhizosphere [34][47].
2.2. From Genome to Gene
The first technique to sequence a gene or genome dates to 1977 by Sanger and Coulson [58], who revolutionized research in biology by contributing to a new viewpoint in molecular biology. This method, among others [59], dominated genetic research up to 2005 [60][61], in which a new method based on automated capillary electrophoresis was developed to improve data knowledge of genes and genome sequences [62], generating pivotal information related to genetics, epigenetics, and transcriptomics. This new technology, named next-generation sequencing (NGS), has opened new scenarios in health, environment, and agriculture-related studies due to the highly accessible low-cost and fast high-throughput sequencing technique [63]. In addition, the capacity to obtain large genomic data sets (Giga base), the scalability, the de novo sequencing and resequencing, the discovery of genomic variants, and molecular markers in crops are other features that distinguish NGS from the older technologies. Moreover, NGS technologies can be applied for TF binding site identification and chromatin alteration studies [59].
Concerning the first aspect (from weed to crops), the study by Guo et al. [54] is an important milestone in the elucidation of the allelopathic mechanisms implemented in barnyardgrass. Since barnyardgrass is among the most widespread weeds in the world, the researchers shed light on the molecular mechanisms underlying its high allelopathic potential. In particular, using genome assembly and annotation (RNA-seq), the researchers identified two gene clusters, involved in DIMBOA and phytoalexin momilactone A biosynthesis, which were activated in response to cocultivation with rice, and detoxification seemed to be the main mechanism conferring an extreme adaptation to the weed [54].
However, the limits of this work could be traced back to the polyploid nature of the species used and the technologies adopted as they do not allow to associate cause–effect in a statistically significant way.
2.3. Plant Breeding in Allelopathy
The main goal of plant breeding is to maintain the quality of life on earth. Improving crops for allelopathy falls within sustainable agriculture, and different strategies, based on genetic variability or transformation, can be adopted to reach this goal. In the first case, natural genetic variability is used to obtain multiple genotypic variants with small phenotypic effects, whereas the genetic transformation forms variants with a significant effect on phenotype. Genetic variability among and within species provides a genetic pool on which to select crop with high allelopathic ability [43], highlighting how improving allelopathy in crops depends on the understanding of the genetic control of these traits. However, as demonstrated by different researchers, allelopathic traits follow a normal distribution, thereby outlining the quantitative nature of the traits and their polygenic control [44][64]. Thus, the approaches used to study and understand multiple traits in plants are quantitative trait loci (QTL) mapping and genome-wide association studies (GWAS). QTL mapping is based on statistical analysis that links phenotypes, and in this context, it is represented by allelopathic traits, with genotypes (chromosome regions) [64]. In recent years, genetic studies have been performed only on crops with considerable economic importance, such as wheat and rice (Table 1). A full-bodied study was performed by Olofsdotter et al. [65], who report some examples in rice. However, it is important not to overlook the environment’s effect on these quantitative traits. Indeed, for these studies, fixed segregant populations are needed for QTL analysis, in which near isogenic lines (NILs), recombinant inbred lines (RILs), and doubled haploid lines (DHLs) represent the most used plant material. Initially, with the lack of current technology, segregation ratios and first-generation molecular markers were taken into consideration, as well as euploid, aneuploid, and substitution lines, to investigate the loci of genes controlling the accumulation of DIMBOA on chromosomes [66]. Yet, QTLs associated with allelopathic traits in rice were also identified using restriction fragment length polymorphism (RFLP) markers in the F2 population from a cross between two contrasting varieties for this trait [67], while Wu et al. [43] identified such QTLs studying the allelopathic effect of wheat on ryegrass using the “equal-compartment agar method”.
High-throughput SNP genotyping was recently adopted to identify QTLs associated with the allelopathic traits in rice [41]. For this purpose, 98 F8 RILs were produced by single-seed descent by crossing a cultivar with high allelopathic potential (Sathi) with a nonallelopathic cultivar (Non-an). On chromosome 8, two QTLs, qlTL-8 and qlSL-8, were detected and were responsible for shoot and root length inhibition, explaining 20 and 15% of the phenotypic variation, respectively. Interestingly, between these QTLs, 31 genes were located [41].
2.4. Microorganism in Allelopathy
Microorganisms colonizing the rhizosphere, mycorrhizosphere, and phyllosphere play a pivotal role in plant health and performance through different mechanisms, including allelopathy [68][69]. There is continuous allelopathic crosstalk between plants and the complex microbial communities in which plant roots secrete a variety of molecules able to shape the rhizosphere microbiota, which in turn produce feedback on the plant [70][71][72]
The role of root-exuded coumarins in shaping the root microbiome clearing the rhizosphere from competing microorganisms to give coumarin-resistant microorganisms a competitive advantage has been recently highlighted and represents a good example of how plants affect soil microorganism communities [73]. Subsequently, Stinglis et al. [74] described the molecular basis of the Arabidopsis thaliana–Pseudomonas simiae WCS417 beneficial model system, in which the plant and the probiotic rhizobacteria closely collaborate to induce the root-specific MYB72 TF and the MYB72-controlled β-glucosidase BGLU42 scopoletin-dependent biosynthesis. The excretion of this metabolite selectively inhibits the soil-borne fungal pathogens Fusarium oxysporum and Verticillium dahliae and promotes the growth of rhizobacteria P. simiae WCS417 and Pseudomonas capeferrum WCS358 responsible for the rhizobacteria-induced systemic resistance (ISR). This molecular collaboration led to plant protection and growth enhancement and improved the niche establishment of the microbial partner as well.
The phenolic acids secreted by allelopathic rice into soil induced the gathering of myxobacteria in the rhizosphere. The latter is responsible for the production of a large number of secondary metabolites with allelopathic activity, among which quercetin, a potential allelochemical deriving from the ferulic acid-induced Myxococcus xanthus cultured medium and playing a role in weed germination and growth suppression. In addition, Escudero-Martinez et al. [75] identified the QRMC-3HS genomic region as the major determinant of the composition of barley rhizosphere microbiota communities. Then, performing a root comparative RNA-seq profiling on the barley lines with contrasting alleles at QRMC-3HS, they identified a nucleotide-binding leucine-rich repeat (NLR) gene among the primary candidate genes.
In their turn, microorganisms produce allelochemicals such as phytohormones (e.g., ABA, auxins, ethylene), volatile organic compounds (e.g., ketones, alcohols, alkanes, terpenoids), quorum sensing molecules (e.g., N-acylhomoserine-lactones, AHL), and antibiotics [76], that can promote plant growth [77], resistance to stress [77][78], induce resistance to diseases, antagonize phytopathogens [79], and control weeds [38][80].
Among rhizosphere microbiota, plant growth-promoting rhizobacteria (PGPR) is a group of beneficial microorganisms (fungi and bacteria) that can promote plant growth by regulating phytohormones synthesis/transport and inducing plant systemic resistance and tolerance through VOCs production. The RNA-seq profiling revealed that VOC treatment affected the expression of 123 genes, among which cell wall modification, auxin induction, stress, and defense response-related genes, with a notable downregulation of several stress-related genes. Furthermore, a transcriptome analysis of the growth-promoting effect of VOCs produced by Microbacterium aurantiacum GX14001 on tobacco (Nicotiana benthamiana) revealed that most of the upregulated genes in response to the bacterium VOCs were involved in plant hormone signal transduction, phenylpropyl biosynthesis, plant–pathogen interaction, and flavonoid biosynthesis pathways. The researchers suggested that plant hormone signal regulation was the way by which GX14001’s VOCs promoted tobacco growth, a suggestion that was validated by further Arabidopsis mutant experiments [81].
Moreover, Berendsen et al. [82] defined the rhizosphere microorganisms as the “plant secondary genome” reporting the impact of the microbe-derived compounds on plant performances. In addition, Rout et al. [83] exposed an interesting perception of the plant microbial genomes as integrated components of the plant genome and highlighted the importance of considering the plant microbiota (all microorganisms) as a plant microbiome (all microbial genomes) that constantly dialogues with the host genes [84]. In addition to the allelochemical substances, a more elaborated communication process, a kind of “molecular allelopathy”, was recently discovered. The “cross-kingdom RNAi” phenomenon is described as a bi-directional communication channel organized by the plant and its associated rhizospheric microorganisms through extracellular vesicles (EVs) carrying miRNAs to induce gene silencing (Figure 1) [85].
References
- Einhellig, F.A. Mechanism of action of allelochemicals in allelopathy. In Allelopathy: Organisms, Processes, and Applications; Inderjit, K.M., Dakshini, M., Einhellig, F.A., Eds.; ACS Symposium Series 582; American Chemical Society: Washington, DC, USA, 1995; pp. 96–116.
- Latif, S.; Chiapusio, G.; Weston, L.A. Allelopathy and the role of allelochemicals in plant defence. Adv. Bot. Res. 2017, 82, 19–54.
- Molisch, H. Der Einfluss einer Pflanze auf die Andere-Allelopathie; Gustav Fisher Verlag: Jena, Germany, 1937.
- Rice, E.L. Allelopathy, 2nd ed.; Academic Press, Inc.: Orlando, FL, USA, 1984.
- Zhang, Z.; Liu, Y.; Yuan, L.; Weber, E.; van Kleunen, M. Effect of allelopathy on plant performance: A meta-analysis. Ecol. Lett. 2021, 24, 348–362.
- Bais, H.P. We are family: Kin recognition in crop plants. New Phytol. 2018, 220, 357–359.
- Biedrzycki, M.L.; Jalany, T.A.; Dudley, S.A.; Bais, H.P. Root exudates mediate kin recognition in plants. Commun. Integr. Biol. 2010, 3, 28–35.
- Crepy, M.A.; Casal, J.J. Photoreceptor-mediated kin recognition in plants. New Phytol. 2015, 205, 329–338.
- Durret, R.; Levin, S. Allelopathy in Spatially Distributed Populations. J. Theor. Biol. 1997, 185, 165–171.
- Inderjit, K.; Callaway, R.M. Experimental designs for the study of allelopathy. Plant Soil 2003, 256, 1–11.
- Inoye, B.; Stinchcombe, J.R. Relationships between ecological interaction modifications and diffuse coevolution: Similarities, differences, and causal links. Oikos 2001, 95, 353–360.
- Guo, L.; Qiu, J.; Li, L.-F.; Lu, B.; Olsen, K.; Fan, L. Genomic clues for crop-weed interactions and evolution. Trends Plant Sci. 2018, 23, 1102–1115.
- Del Moral, R.; Willis, R.J.; Ashton, D.H. Suppression of coastal heath vegetation by Eucalyptus baxteri. Aust. J. Bot. 1978, 26, 203–219.
- Fischer, N.H.; Williamson, G.B.; Weidenhamer, J.D.; Richardson, D.R. In search of allelopathy in the Florida scrub: The role of terpenoids. J. Chem. Ecol. 1994, 20, 1355–1380.
- Katz, D.A.; Sneh, B.; Friedman, J. The allelopathic potential of Coridothymus capitatus L. (Labiatae). Preliminary studies on the roles of the shrub in the inhibition of annuals germination and/or to promote allelopathically active actinomycetes. Plant Soil 1987, 98, 53–66.
- Weidenhamer, J.D.; Romeo, J.T. Allelopathic properties of Polygonella myriphylla: Field evidence and bioassay. J. Chem. Ecol. 1989, 15, 1957–1970.
- Bais, H.P.; Vepachedu, R.; Gilroy, S.; Callaway, R.M.; Vivanco, J.M. Allelopathy and exotic plant invasion: From molecules and genes to species interactions. Science 2003, 301, 1377–1380.
- Schenk, H.J.; Callaway, R.M.; Mahall, B.E. Spatial Root Segregation: Are Plants Territorial? Adv. Ecol. Res. 1999, 28, 145–180.
- Rizvi, S.G.H.; Rizvi, V. Allelopathy: Basic and Applied Aspects; Chapman and Hall: London, UK, 1992.
- Bais, H.P.; Walker, T.S.; Stermitz, F.R.; Hufbauer, R.A.; Vivanco, J.M. Enantiomeric-dependent phytotoxic and antimicrobial activity of (±)-catechin. A rhizosecreted racemic mixture from spotted knapweed. Plant Physiol. 2002, 128, 1173–1179.
- Macías, F.A.; Mejías, F.J.R.; Molinillo, J.M.G. Recent advances in allelopathy for weed control: From knowledge to applications. Pest. Manag. Sci. 2019, 75, 2413–2436.
- Lupini, A.; Sorgonà, A.; Princi, M.P.; Sunseri, F.; Abenavoli, M.R. Morphological and physiological effects of trans-cinnamic acid and its hydroxylated derivatives on maize root types. Plant Growth Regul. 2016, 78, 263–273.
- Politycka, B. Free and glucosylated phenolics, phenol-beta-glucosyltransferase activity and membrane permeability in cucumber roots affected by derivatives of cinnamic and benzoic acid. Acta Physiol. Plantarum 1997, 19, 311–317.
- Cruz, O.R.; Anaya, A.L.; Hernandez-Bautista, B.E. Effects of allelochemical stress produced by sicyosdeppei on seedling root ultrastructure of Phaseolous vulgaris and Cucubita ficifolia. J. Chem. Ecol. 1998, 24, 2039–2057.
- Patterson, D.T. Effects of allelopathic chemicals on growth and physiological response of soybean (Glycine max). Weed Sci. 1981, 29, 53–58.
- Araniti, F.; Bruno, L.; Sunseri, F.; Pacenza, M.; Forgione, I.; Bitonti, M.B.; Abenavoli, M.R. The allelochemical farnesene affects Arabidopsis thaliana root meristem altering auxin distribution. Plant Physiol. Bioch. 2017, 121, 14–20.
- Yazaki, K.; Arimura, G.; Ohnishi, T. “Hidden” terpenoids in plants: Their biosynthesis, localization and ecological roles. Plant Cell Physiol. 2017, 58, 1615–1621.
- Gershenzon, J.; Dudareva, N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007, 3, 408–414.
- Chou, C.C. Introduction to allelopathy. In Allelopathy: A Physiological Process with Ecological Implications; Reigosa, M.J., Pedrol, N., González, L., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 1–9.
- Zimdahl, R.L. Weed–Crop Competition: A Review, 2nd ed.; Blackwell: Hoboken, NJ, USA, 2004.
- MacLaren, C.; Storkey, J.; Menagat, A.; Metcalfe, H.; Dehnen-Schmutz, K. An ecological future for weed science to sustain crop production and the environment. A review. Agron. Sustain. Dev. 2020, 40, 24.
- Duke, S.O.; Baerson, S.R.; Pan, Z.; Kagan, I.A.; Sanchez-Moreiras, A.; Reigosa, M.J.; Pedrol, N.; Schulz, M. Genomic Approaches to Understanding Allelochemical Effects on Plants. In Allelopathy in Sustainable Agriculture and Forestry; Zeng, R.S., Mallik, A.U., Luo, S.M., Eds.; Springer: New York, NY, USA, 2008.
- Yuan, Y.; Lee, H.T.; Hu, H.; Scheben, A.; Edwards, D. Single-Cell genomics analysis in plants. Genes 2018, 9, 50.
- He, H.Q.; Lin, W.X.; Liang, Y.Y.; Song, B.Q.; Ke, Y.Q.; Guo, Y.C.; Liang, K.J. Analyzing the molecular mechanism of crop allelopathy by using differential proteomics. Acta Ecol. Sin. 2005, 25, 3141–3146.
- Chi, W.-C.; Chen, Y.-A.; Hsiung, Y.-C.; Fu, S.-F.; Chou, C.-H.; Trinh, N.N.; Chen, Y.-C.; Huang, H.-J. Autotoxicity mechanism of Oryza sativa: Transcriptome response in rice roots exposed to ferulic acid. BMC Genom. 2013, 14, 351.
- Cheng, F.; Cheng, Z.H.; Meng, H.W. Transcriptomic insights into the allelopathic effects of the garlic allelochemical diallyl disulfide on tomato roots. Sci. Rep. 2016, 6, 38902.
- Zhang, Q.; Zheng, X.-Y.; Lin, S.-X.; Gu, C.-Z.; Li, L.; Li, J.-Y.; Fang, C.-X.; He, H.-B. Transcriptome analysis reveals that barnyard grass exudates increase the allelopathic potential of allelopathic and non-allelopathic rice (Oryza sativa) accessions. Rice 2019, 12, 30.
- Li, L.-L.; Zhao, H.-H.; Kong, C.-H. (-)-Loliolide, the most ubiquitous lactone, is involved in barnyard grass-induced rice allelopathy. J. Exp. Bot. 2020, 71, 1540–1550.
- Yang, Y.; Zhang, Z.; Li, R.; Yi, Y.; Yang, H.; Wang, C.; Wang, Z.; Liu, Y. RgC3H involves in the biosynthesis of allelopathic phenolic acids and alters their release amount in Rehmannia glutinosa roots. Plants 2020, 9, 567.
- Horvath, D.P.; Hansen, S.A.; Moriles-Miller, J.P.; Pierik, R.; Yan, C.; Clay, D.E.; Scheffler, B.; Clay, S.A. RNAseq reveals weed-induced PIF3-like as candidate target to manipulate weed stress response in soybean. New Phytol. 2015, 207, 196–210.
- Chung, I.-M.; Ham, T.-H.; Cho, G.-W.; Kwon, S.-W.; Lee, Y.; Seo, J.; An, Y.-J.; Kim, S.-Y.; Lee, J. Study of quantitative trait loci (QTLs) associated with allelopathic trait in rice. Genes 2020, 11, 479.
- Zhang, Q.; Zhang, Q.; Lin, S.; Wang, P.; Li, J.; Wang, H.; He, H. Dynamic analysis on weed inhibition and phenolic acids of allelopathic rice in field test. Arch. Agron. Soil Sci. 2020, 67, 1809–1821.
- Wu, H.; Pratley, J.; Ma, W.; Haig, T. Quantitative trait loci and molecular markers associated with wheat allelopathy. Theor. Appl. Genet. 2003, 107, 1477–1481.
- Zeng, D.; Qian, Q.; Teng, S.; Dong, G.; Fujimoto, H.; Yasufumi, K.; Zhu, L. Genetic analysis of rice allelopathy. Chin. Sci. Bull. 2003, 48, 265–268.
- Quader, M.; Daggard, G.; Barrow, R.; Walker, S.; Sutherland, M.W. Allelopathy, Dimboa production and genetic variability in accessions of Triticum speltoides. J. Chem. Ecol. 2001, 27, 747–760.
- Fang, C.; Zhuang, Y.; Xu, T.; Li, Y.; Li, Y.; Lin, W. Changes in rice allelopathy and rhizosphere microflora by inhibiting rice phenylalanine ammonialyase gene expression. J. Chem. Ecol. 2013, 39, 204–212.
- Xu, M.; Galhano, R.; Wiemann, P.; Bueno, E.; Tiernan, M.; Wu, W.; Peters, R.J. Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa). New Phytol. 2012, 193, 570–575.
- Zhang, Q.; Lin, S.; Zhang, Q.; Wang, P.; Wang, H.; He, H. Comparative analysis on metabolites and gene expression difference of various allelopathic potential rice under weed stress. Weed Res. 2022; in press.
- Shehzad, T.; Okuno, K. Genetic analysis of QTLs controlling allelopathic characteristics in sorghum. PLoS ONE 2020, 15, e0235896.
- Fang, C.; Yang, L.; Chen, W.; Li, L.; Zhang, P.; Li, Y.; He, H.; Lin, W. MYB57 transcriptionally regulates MAPK11 to interact with PAL2;3 and modulate rice allelopathy. J. Exp. Bot. 2020, 71, 2127–2142.
- Sultana, M.H.; Liu, F.; Alamin, M.; Mao, L.; Jia, L.; Chen, H.; Wu, D.; Wang, Y.; Fu, F.; Wu, S.; et al. Gene modules co-regulated with biosynthetic gene custers for allelopathy between rice and bernyargrass. Int. J. Mol. Sci. 2019, 20, 3846.
- Zhang, H.; Rutherford, S.; Qi, S.; Huang, P.; Dai, Z. Transcriptome profiling of Arabidopsis thaliana roots in response to allelopathic effects of Conyza canadensis. Ecotoxicology 2022, 31, 53–63.
- Li, J.; Chen, L.; Chen, Q.; Miao, Y.; Peng, Z.; Huang, B.; Guo, L.; Liu, D.; Du, H. Allelopathic effect of Artemisia argyi on the germination and growth of various weeds. Sci. Rep. 2021, 11, 4303.
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L.; et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 2017, 8, 1031.
- Gierl, A.; Frey, M. Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 2001, 213, 493–498.
- Frey, M.; Chomet, P.; Glawischnig, E.; Stettner, C.; Grün, S.; Winklmair, A.; Eisenreich, W.; Bacher, A.; Meerley, R.B.; Briggs, S.P.; et al. Analysis of a chemical plant defense mechanism in grasses. Science 1997, 277, 696–699.
- Song, B.; Xiong, J.; Fang, C.; Qiu, L.; Lin, R.; Liang, Y.; Lin, W. Allelopathic enhancement and differential gene expression in rice under low nitrogen treatment. J. Chem. Ecol. 2008, 34, 688–695.
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467.
- Afzal, F.; Gul, A.; Kazi, A.M. Next-generation sequencing technologies and plant improvement. In Plant Omics: Trends and Applications, 1st ed.; Hakeem, K.R., Tombuloğlu, H., Tombuloğlu, G., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 271–294.
- Maxam, A.M.; Gilbert, W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 1977, 74, 560–564.
- Morozova, O.; Marra, M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics 2008, 92, 255–264.
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18.
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2006, 437, 376–380.
- Asins, M.J.; Bernet, G.P.; Villalta, I.; Carbonell, E.A. QTL Analysis in Plant Breeding. In Molecular Techniques in Crop Improvement; Jain, S., Brar, D., Eds.; Springer: Dordrecht, The Netherlands, 2010.
- Olofsdotter, M.; Jensen, L.B.; Courtois, B. Improving crop competitive ability using allelopathy—An example from rice. Plant Breed. 2002, 121, 1–9.
- Niemeyer, H.M.; Jerez, J.M. Chromosomal location of genes for hydroxamic acid accumulation in wheat using wheat aneuploids and wheat substitution lines. Heredity 1997, 79, 10–14.
- Ebana, K.; Yan, W.; Dilday, R.H.; Namai, H.; Okuno, K. Analysis of QTL Associated with the Allelopathic Effect of Rice Using Water-soluble Extracts. Breed. Sci. 2001, 51, 47–51.
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590.
- Müller, H.; Berg, C.; Landa, B.B.; Auerbach, A.; Moissl-Eichinger, C.; Berg, G. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize mediterranean olive trees. Front. Microbiol. 2015, 6, 138.
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal Biodiversity and Their Role in Soil Health. Front. Microbiol. 2018, 9, 707.
- Barrera, S.E.; Sarango-Flóres, S.W.; Montenegro-Gómez, S.P. The phyllosphere microbiome and its potential application in horticultural crops. A review. Rev. Colomb. Cienc. Hortic. 2019, 13, 384–396.
- Ding, L.-J.; Cui, H.-L.; Nie, S.-A.; Long, X.-E.; Duan, G.-L.; Zhu, Y.-G. Microbiomes inhabiting rice roots and rhizosphere. FEMS Microbiol. Ecol. 2019, 95, fiz040.
- Lundberg, D.S.; Teixeira, P.J. Root-exuded coumarin shapes the root microbiome. Proc. Natl. Acad. Sci. USA 2018, 115, 5629–5631.
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222.
- Escudero-Martinez, C.; Coulter, M.; Alegria Terrazas, R.; Foito, A.; Kapadia, R.; Pietrangelo, L.; Maver, M.; Sharma, R.; Aprile, A.; Morris, J.; et al. Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nat. Commun. 2022, 13, 3443.
- Müller, C.A.; Obermeier, M.M.; Berg, G. Bioprospecting plant-associated microbiomes. J. Biotechnol. 2016, 235, 171–180.
- Egamberdieva, D.; Wirth, S.; Alqarawi, A.A.; Elsayed, F.A.A.; Hashem, A. Phytohormones and Beneficial Microbes: Essential Components for Plants to Balance Stress and Fitness. Front. Microbiol. 2017, 8, 2104.
- Ho, N.-G.; Mathew, D.C.; Huang, C.-C. Plant-Microbe Ecology: Interactions of Plants and Symbiotic Microbial Communities. In Plant Ecology-Traditional Approaches to Recent Trends; IntechOpen: London, UK, 2017.
- Yi, H.-S.; Yang, J.W.; Ryu, C.-M. ISR meets SAR outside: Additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spot in field-grown pepper. Front. Plant Sci. 2013, 4, 122.
- Abbas, T.; Zahir, Z.A.; Naveed, M.; Abbas, S.; Alwahibi, M.S.; Elshikh, M.S.; Adnan, M. Large Scale Screening of Rhizospheric Allelopathic Bacteria and Their Potential for the Biocontrol of Wheat-Associated Weeds. Agronomy 2020, 10, 1469.
- Gao, Y.; Feng, J.; Wu, J.; Wang, K.; Wu, S.; Liu, H.; Jiang, M. Transcriptome analysis of the growth-promoting effect of volatile organic compounds produced by Microbacterium aurantiacum GX14001 on tobacco (Nicotiana benthamiana). BMC Plant Biol. 2022, 22, 208.
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486.
- Rout, M.E.; Southworth, D. The root microbiome influences scales from molecules to ecosystems: The unseen majority. Am. J. Bot. 2013, 100, 1689–1691.
- Bever, J.D. Feedback between plants and their soil communities in an old field community. Ecology 1994, 75, 1965–1977.
- Baldrich, P.; Rutter, B.D.; Karimi, H.Z.; Podicheti, R.; Meyers, B.C.; Innes, R.W. Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-nucleotide “tiny” RNAs. Plant Cell 2019, 31, 315–324.
More
Information
Subjects:
Agronomy
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
27 Sep 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




