
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yingbin Xue | -- | 2328 | 2022-09-09 11:00:30 | | | |
| 2 | Rita Xu | -3 word(s) | 2325 | 2022-09-09 11:18:43 | | |
Video Upload Options
Low phosphate (Pi) availability in soils severely limits crop growth and production. Plants have evolved to have numerous physiological and molecular adaptive mechanisms to cope with Pi starvation. The release of Pi from membrane phospholipids is considered to improve plant phosphorus (P) utilization efficiency in response to Pi starvation and accompanies membrane lipid remodeling. Researchers summarize recent discoveries related to this topic and the molecular basis of membrane phospholipid alteration in response to Pi depletion in plants at different subcellular levels. These findings will help to further elucidate the molecular mechanisms underlying plant adaptation to Pi starvation and thus help to develop crop cultivars with high P utilization efficiency.
1. Introduction
2. Phospholipids’ Degradation in Response to P Deficiency
2.1. Phospholipase C Pathway
2.2. Phospholipase D and Phosphatidic Acid Phosphatase Pathway
2.3. Lipid Acyl Hydrolase and Glycerophosphodiester Phosphodiesterase Pathway
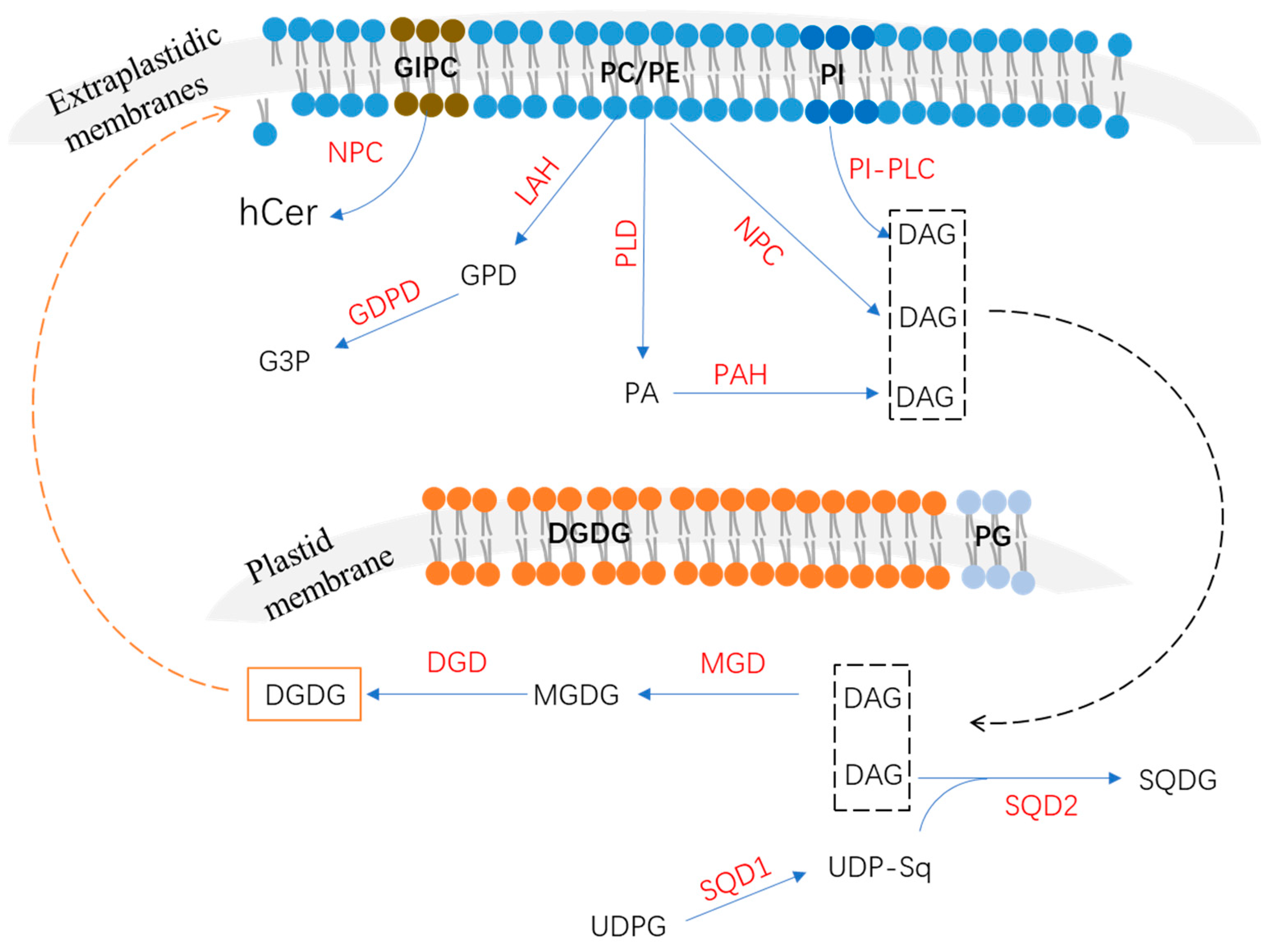
3. Non-Phosphorus Lipid Biosynthesis
3.1. Biosynthesis of Monogalactosyldiacylglycerol and Digalactosyldiacylglycerol
3.2. Biosynthesis of Sulfoquinovosyldiacylglycerol
References
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66.
- Lambers, H. Phosphorus acquisition and utilization in plants. Annu. Rev. Plant Biol. 2022, 73, 17–42.
- Hallama, M.; Pekrun, C.; Lambers, H.; Kandeler, E. Hidden miners–the roles of cover crops and soil microorganisms in phosphorus cycling through agroecosystems. Plant Soil 2019, 434, 7–45.
- Veneklaas, E.J.; Lambers, H.; Bragg, J.; Finnegan, P.M.; Lovelock, C.E.; Plaxton, W.C.; Price, C.A.; Scheible, W.R.; Shane, M.W.; White, P.J.; et al. Opportunities for improving phosphorus-use efficiency in crop plants. N. Phytol. 2012, 195, 306–320.
- Wang, L.; Liu, D. Functions and regulation of phosphate starvationinduced secreted acid phosphatases in higher plants. Plant Sci. 2018, 271, 108–116.
- Puga, M.I.; Rojas-Triana, M.; de Lorenzo, L.; Leyva, A.; Rubio, V.; Paz-Ares, J. Novel signals in the regulation of Pi starvation responses in plants: Facts and promises. Curr. Opin. Plant Biol. 2017, 39, 40–49.
- Gutiérrez-Alanís, D.; Ojeda-Rivera, J.O.; Yong-Villalobos, L.; Cárdenas-Torres, L.; Herrera-Estrella, L. Adaptation to phosphate scarcity: Tips from Arabidopsis roots. Trends Plant Sci. 2018, 23, 721–730.
- Nakamura, Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 2013, 52, 43–50.
- Su, W.; Raza, A.; Gao, A.; Zeng, L.; Lv, Y.; Ding, X.; Cheng, Y.; Zou, X. Plant lipid phosphate phosphatases: Current advances and future outlooks. Crit. Rev. Biotechnol. 2022, 17, 1–9.
- Verma, L.; Kohli, P.S.; Maurya, K.; Abhijith, K.B.; Thakur, J.K.; Giri, J. Specific galactolipids species correlate with rice genotypic variability for phosphate utilization efficiency. Plant Physiol. Biochem. 2021, 168, 105–115.
- Nakamura, Y.; Koizumi, R.; Shui, G.; Shimojima, M.; Wenk, M.R.; Ito, T.; Ohta, H. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. USA 2009, 106, 20978–20983.
- Pfaff, J.; Denton, A.K.; Usadel, B.; Pfaff, C. Phosphate starvation causes different stress responses in the lipid metabolism of tomato leaves and roots. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 2020, 1865, 158763.
- Joyard, J.; Teyssier, E.; Miege, C.; Berny-Seigneurin, D.; Marechal, E.; Block, M.A.; Dorne, A.J.; Rolland, N.; Ajlani, G.; Douce, R. The biochemical machinery of plastid envelope membranes. Plant Physiol. 1998, 118, 715–723.
- Singh, A.; Bhatnagar, N.; Pandey, A.; Pandey, G.K. Plant phospholipase C family: Regulation and functional role in lipid signaling. Cell Calcium. 2015, 58, 139–146.
- Sagar, S.; Singh, A. Emerging role of phospholipase C mediated lipid signaling in abiotic stress tolerance and development in plants. Plant Cell Rep. 2021, 40, 2123–2133.
- Mueller-Roeber, B.; Pical, C. Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 2002, 130, 22–46.
- Hunt, L.; Otterhag, L.; Lee, J.C.; Lasheen, T.; Hunt, J.; Seki, M.; Shinozaki, K.; Sommarin, M.; Gilmour, D.J.; Pical, C.; et al. Gene-specific expression and calcium activation of Arabidopsis thaliana phospholipase C isoforms. N. Phytol. 2004, 162, 643–654.
- Tasma, I.M.; Brendel, V.; Whitham, S.A.; Bhattacharyya, M.K. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 2008, 46, 627–637.
- Kanehara, K.; Yu, C.Y.; Cho, Y.; Cheong, W.F.; Torta, F.; Shui, G.; Wenk, M.R.; Nakamura, Y. Arabidopsis AtPLC2 is a primary phosphoinositide-specific phospholipase c in phosphoinositide metabolism and the endoplasmic reticulum stress response. PLoS Genet. 2015, 11, e1005511.
- Nakamura, Y.; Awai, K.; Masuda, T.; Yoshioka, Y.; Takamiya, K.; Ohta, H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 2005, 280, 7469–7476.
- Su, Y.; Li, M.; Guo, L.; Wang, X. Different effects of phospholipase Dζ2 and non-specific phospholipase C4 on lipid remodeling and root hair growth in Arabidopsis response to phosphate deficiency. Plant J. 2018, 94, 315–326.
- Peters, C.; Li, M.; Narasimhan, R.; Roth, M.; Welti, R.; Wang, X. Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell. 2010, 22, 2642–2659.
- Yang, B.; Li, M.; Phillips, A.; Li, L.; Ali, U.; Li, Q.; Lu, S.; Hong, Y.; Wang, X.; Guo, L. Nonspecific phospholipase C4 hydrolyzes phosphosphingolipids and sustains plant root growth during phosphate deficiency. Plant Cell. 2021, 33, 766–780.
- Yang, B.; Zhang, K.; Jin, X.; Yan, J.; Lu, S.; Shen, Q.; Guo, L.; Hong, Y.; Wang, X.; Guo, L. Acylation of non-specific phospholipase C4 determines its function in plant response to phosphate deficiency. Plant J. 2021, 106, 1647–1659.
- Peters, C.; Kim, S.C.; Devaiah, S.; Li, M.; Wang, X. Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 2014, 37, 2002–2013.
- Wang, X. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 2005, 139, 566–573.
- Ali, U.; Lu, S.; Fadlalla, T.; Iqbal, S.; Yue, H.; Yang, B.; Hong, Y.; Wang, X.; Guo, L. The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses. Prog. Lipid Res. 2022, 86, 101158.
- Cruz-Ramírez, A.; Oropeza-Aburto, A.; Razo-Hernández, F.; Ramírez-Chávez, E.; Herrera-Estrella, L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2006, 103, 6765–6770.
- Yamaryo, Y.; Dubots, E.; Albrieux, C.; Baldan, B.; Block, M.A. Phosphate availability affects the tonoplast localization of PLDzeta2, an Arabidopsis thaliana phospholipase D. FEBS Lett. 2008, 582, 685–690.
- Lin, D.L.; Yao, H.Y.; Jia, L.H.; Tan, J.F.; Xu, Z.H.; Zheng, W.M.; Xue, H.W. Phospholipase D-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. N. Phytol. 2020, 226, 142–155.
- Carman, G.M.; Han, G.S. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 2006, 31, 694–699.
- Eastmond, P.J.; Quettier, A.L.; Kroon, J.T.; Craddock, C.; Adams, N.; Slabas, A.R. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell. 2010, 22, 2796–2811.
- Brindley, D.N.; Pilquil, C. Lipid phosphate phosphatases and signaling. J. Lipid Res. 2009, 50, S225–S230.
- Nakamura, Y.; Tsuchiya, M.; Ohta, H. Plastidic phosphatidic acid phosphatases identified in a distinct subfamily of lipid phosphate phosphatases with prokaryotic origin. J. Biol. Chem. 2007, 282, 29013–29021.
- Cheng, Y.; Zhou, W.; El Sheery, N.I.; Peters, C.; Li, M.; Wang, X.; Huang, J. Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 2011, 66, 781–795.
- Morcuende, R.; Bari, R.; Gibon, Y.; Zheng, W.; Pant, B.D.; Bläsing, O.; Usadel, B.; Czechowski, T.; Udvardi, M.K.; Stitt, M.; et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007, 30, 85–112.
- Ngo, A.H.; Nakamura, Y. Phosphate starvation-inducible GLYCEROPHOSPHODIESTER PHOSPHODIESTERASE6 is involved in Arabidopsis root growth. J. Exp. Bot. 2022, 73, 2995–3003.
- Mehra, P.; Giri, J. Rice and chickpea GDPDs are preferentially influenced by low phosphate and CaGDPD1 encodes an active glycerophosphodiester phosphodiesterase enzyme. Plant Cell Rep. 2016, 35, 1699–1717.
- Mehra, P.; Pandey, B.K.; Verma, L.; Giri, J. A novel glycerophosphodiester phosphodiesterase improves phosphate deficiency tolerance in rice. Plant Cell Environ. 2019, 42, 1167–1179.
- Dissanayaka, D.M.S.B.; Ghahremani, M.; Siebers, M.; Wasaki, J.; Plaxton, W.C. Recent insights into the metabolic adaptations of phosphorus-deprived plants. J. Exp. Bot. 2021, 72, 199–223.
- Shimojima, M. Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 2011, 50, 234–239.
- Awai, K.; Maréchal, E.; Block, M.A.; Brun, D.; Masuda, T.; Shimada, H.; Takamiya, K.; Ohta, H.; Joyard, J. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2001, 98, 10960–10965.
- Kobayashi, K.; Awai, K.; Takamiya, K.; Ohta, H. Arabidopsis type B monogalactosyldiacylglycerol synthase genes are expressed during pollen tube growth and induced by phosphate starvation. Plant Physiol. 2004, 134, 640–648.
- Kobayashi, K.; Awai, K.; Nakamura, M.; Nagatani, A.; Masuda, T.; Ohta, H. Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 2009, 57, 322–331.
- Verma, L.; Bhadouria, J.; Rupam, B.K.; Singh, S.; Panchal, P.; Bhatia, C.; Eastmond, P.J.; Giri, J. Monogalactosyl Diacylglycerol Synthase 3 (OsMGD3) affects phosphate utilization and acquisition in rice. J. Exp. Bot. 2022, 73, 5033–5051.
- Basnet, R.; Zhang, J.; Hussain, N.; Shu, Q. Characterization and mutational analysis of a monogalactosyldiacylglycerol synthase gene OsMGD2 in rice. Front. Plant Sci. 2019, 10, 992.
- Kelly, A.A.; Froehlich, J.E.; Dörmann, P. Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell. 2003, 15, 2694–7206.
- Kelly, A.A.; Dörmann, P. DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 2002, 277, 1166–1173.
- Fu, B.; Brown, C.; Mäler, L. Expression and purification of DGD2, a chloroplast outer membrane-associated glycosyltransferase for galactolipid synthesis. Biochemistry 2020, 59, 999–1009.
- Härtel, H.; Dormann, P.; Benning, C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 10649–10654.
- Sanda, S.; Leustek, T.; Theisen, M.J.; Garavito, R.M.; Benning, C. Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 2001, 276, 3941–3946.
- Yu, B.; Xu, C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737.
- Okazaki, Y.; Shimojima, M.; Sawada, Y.; Toyooka, K.; Narisawa, T.; Mochida, K.; Tanaka, H.; Matsuda, F.; Hirai, A.; Hirai, M.Y.; et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 2009, 21, 892–909.
- Sun, Y.; Jain, A.; Xue, Y.; Wang, X.; Zhao, G.; Liu, L.; Hu, Z.; Hu, S.; Shen, X.; Liu, X.; et al. OsSQD1 at the crossroads of phosphate and sulfur metabolism affects plant morphology and lipid composition in response to phosphate deprivation. Plant Cell Environ. 2020, 43, 1669–1690.




