
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Roberta Maestro | -- | 1045 | 2022-09-05 11:53:06 | | | |
| 2 | Jessie Wu | + 10 word(s) | 1055 | 2022-09-08 06:07:32 | | | | |
| 3 | Jessie Wu | -10 word(s) | 1045 | 2022-09-13 05:06:43 | | | | |
| 4 | Jessie Wu | Meta information modification | 1045 | 2022-09-13 05:36:14 | | | | |
| 5 | Jessie Wu | Meta information modification | 1045 | 2022-09-13 05:39:36 | | |
Video Upload Options
SMARCB1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1) is a key component of the SWI/SNF (SWItch/Sucrose Non-Fermentable) chromatin remodeling complexes. Functional inactivation of SMARCB1 is the only recurrent genetic alteration reported in so far in Epithelioid Sarcoma (ES), a very rare and aggressive mesenchymal tumor of unclear origin and uncertain lineage characterized by a prevalent epithelioid morphology.
1. Introduction
2. SMARCB1 inactivation in the pathogenesis of Epithelioid Sarcoma
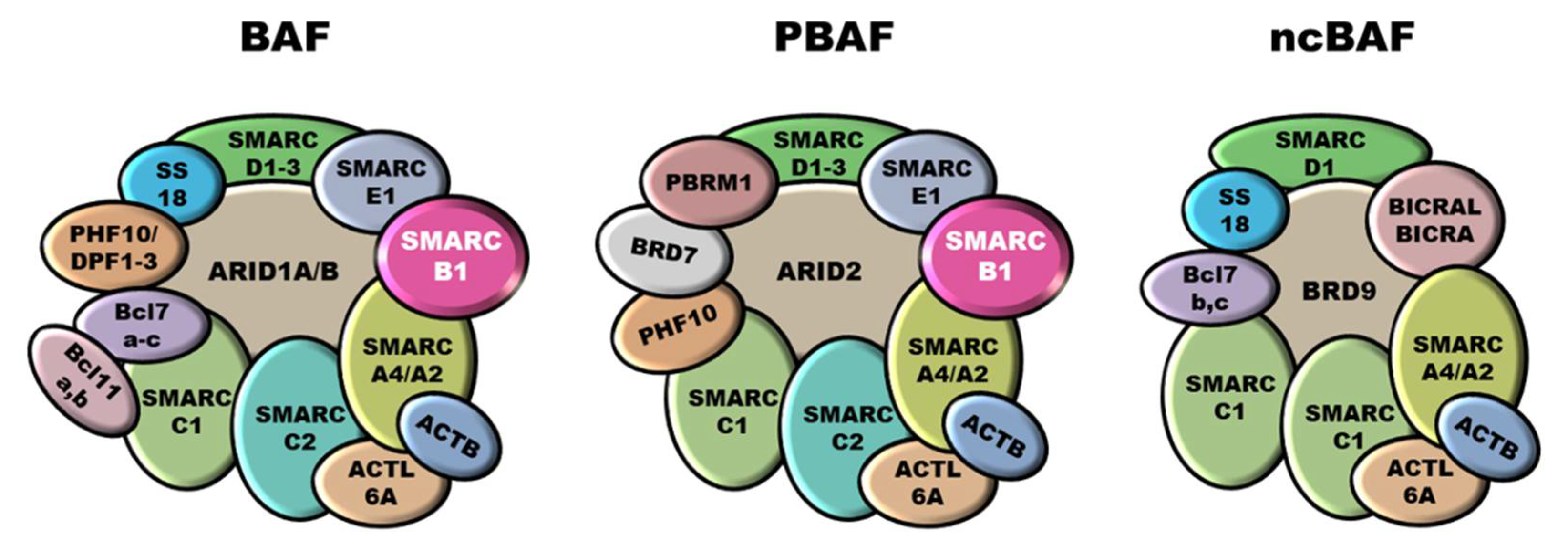
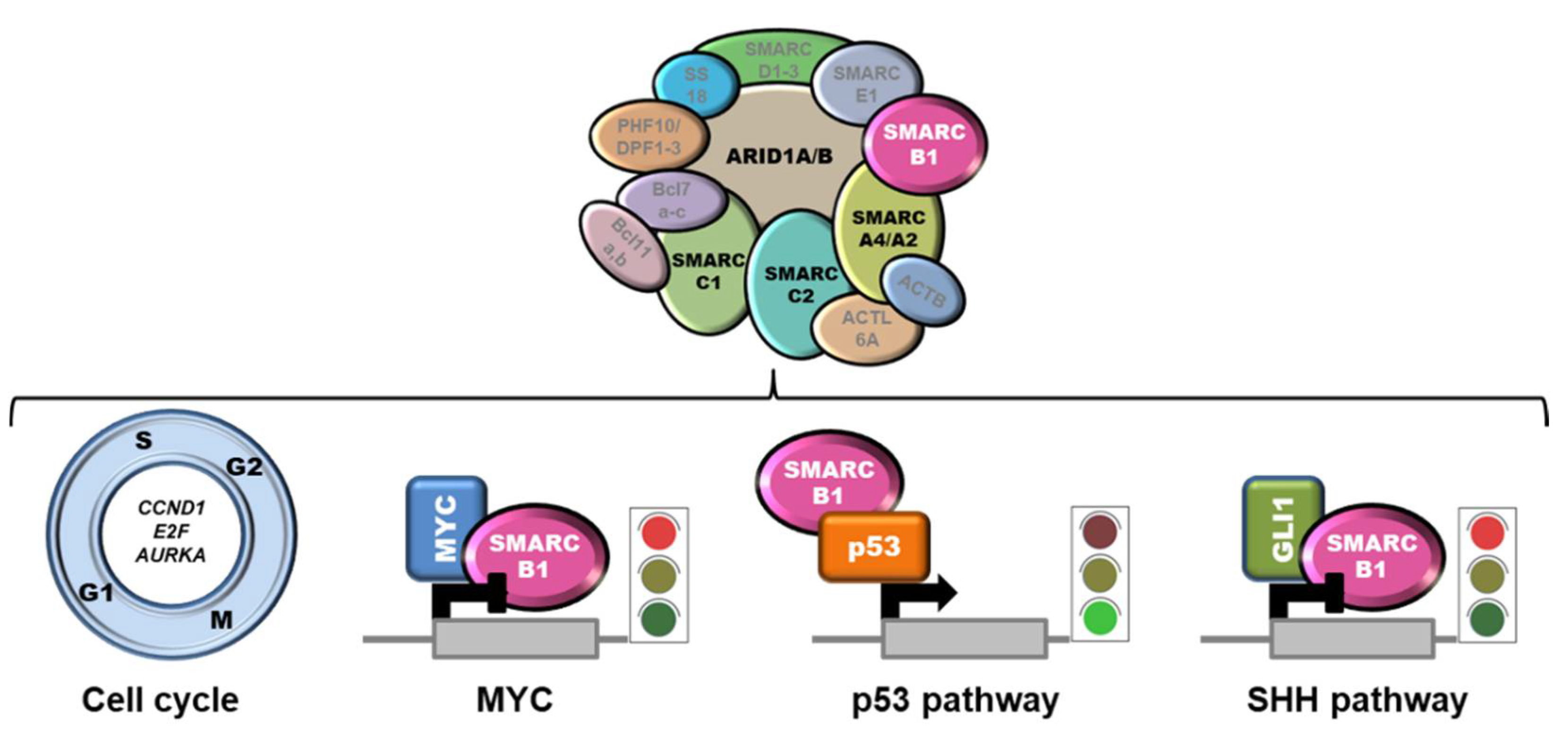
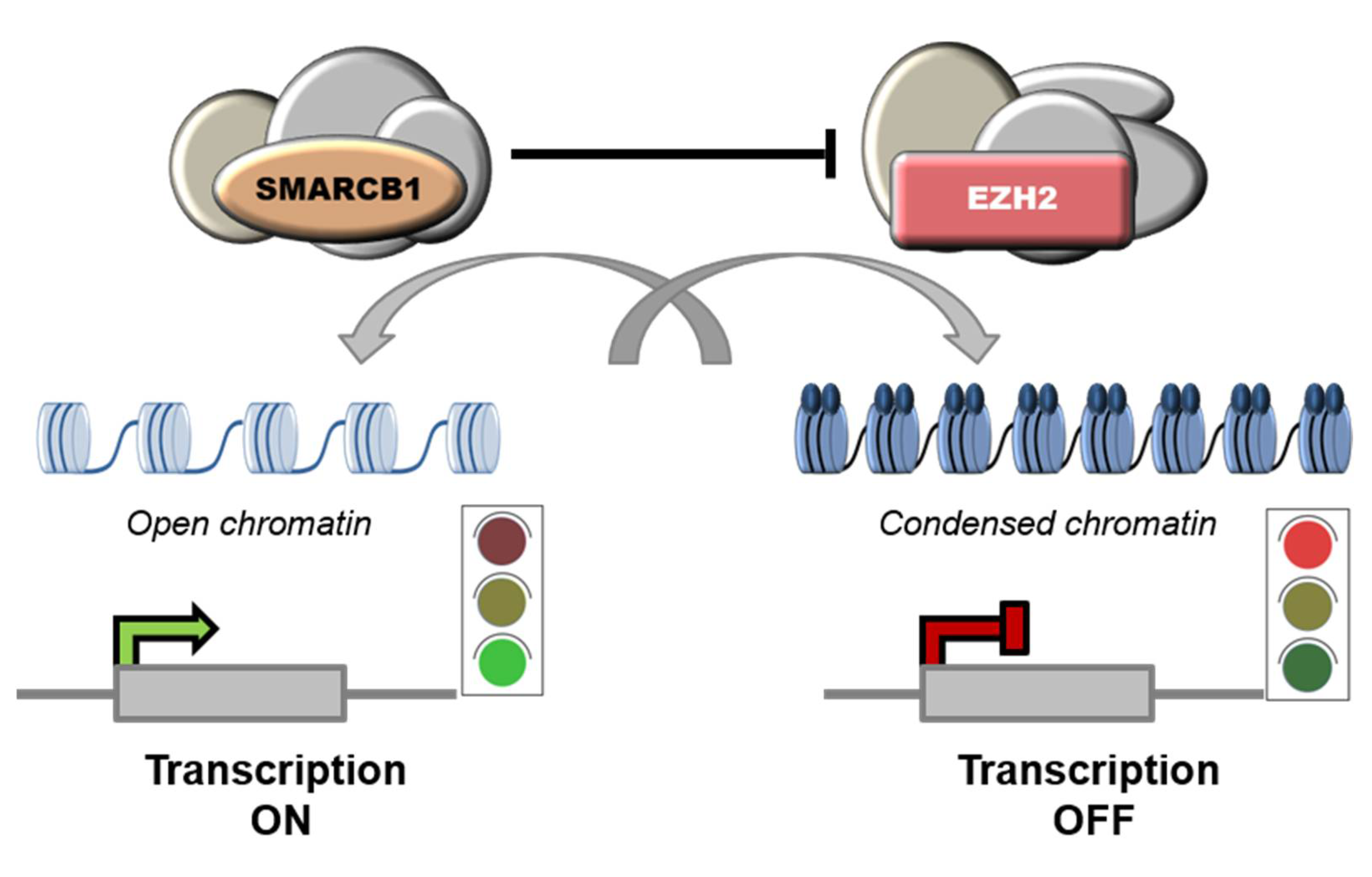
SMARCB1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1), a.k.a. INI1 (Integrase Interactor 1), is a subunit of the mammalian SWI/SNF (SWItch/Sucrose Non-Fermentable) ATP-dependent chromatin remodeling complexes, also known as BAF (BRG1/BRM-associated factor) complexes. These complexes are central regulators of nucleosome remodeling and are implicated in different biological processes, including cell cycle control and maintenance of genomic stability [14][15][16]. Mammalian SWI/SNF complexes are classified into three subgroups: canonical BAF (cBAF), polybromo-associated BAF (PBAF), and non-canonical BAF (ncBAF), also called GLTSCR1 or GLTSCR1L-containing and BRD9-containing (GBAF) complexes [17][18] (Figure 1). SMARCB1 participates only into cBAF and PBAF complexes [19][20]. Notably, SWI/SNF subunits are mutated in over 20% of all human cancers [16][21]. In particular, SMARCB1 inactivation is implicated in the pathobiology of ES, malignant rhabdoid tumors (MRT), atypical teratoid/rhabdoid tumors (AT/RT) of the central nervous system, malignant peripheral nerve sheath tumors (MPNST), myoepithelial neoplasms and renal medullary carcinomas (RMC) [22][23][24][25].
References
- Frezza, A.M.; Botta, L.; Pasquali, S.; Stacchiotti, S.; Gronchi, A.; Casali, P.G.; Trama, A.; Rarecarenet, W.G. An Epidemiological Insight into Epithelioid Sarcoma (ES): The Open Issue of Distal-Type (DES) versus Proximal-Type (PES). Ann. Oncol. 2017, 28, v525.
- Casanova, M.; Ferrari, A.; Collini, P.; Bisogno, G.; Alaggio, R.; Cecchetto, G.; Gronchi, A.; Meazza, C.; Garaventa, A.; Di Cataldo, A.; et al. Epithelioid Sarcoma in Children and Adolescents: A Report from the Italian Soft Tissue Sarcoma Committee. Cancer 2006, 106, 708–717.
- Jawad, M.U.; Extein, J.; Min, E.S.; Scully, S.P. Prognostic Factors for Survival in Patients with Epithelioid Sarcoma: 441 Cases from the SEER Database. Clin. Orthop. 2009, 467, 2939–2948.
- Chase, D.R.; Enzinger, F.M. Epithelioid Sarcoma. Diagnosis, Prognostic Indicators, and Treatment. Am. J. Surg. Pathol. 1985, 9, 241–263.
- Ross, H.M.; Lewis, J.J.; Woodruff, J.M.; Brennan, M.F. Epithelioid Sarcoma: Clinical Behavior and Prognostic Factors of Survival. Ann. Surg. Oncol. 1997, 4, 491–495.
- Callister, M.D.; Ballo, M.T.; Pisters, P.W.; Patel, S.R.; Feig, B.W.; Pollock, R.E.; Benjamin, R.S.; Zagars, G.K. Epithelioid Sarcoma: Results of Conservative Surgery and Radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2001, 51, 384–391.
- Baratti, D.; Pennacchioli, E.; Casali, P.G.; Bertulli, R.; Lozza, L.; Olmi, P.; Collini, P.; Radaelli, S.; Fiore, M.; Gronchi, A. Epithelioid Sarcoma: Prognostic Factors and Survival in a Series of Patients Treated at a Single Institution. Ann. Surg. Oncol. 2007, 14, 3542–3551.
- Wolf, P.S.; Flum, D.R.; Tanas, M.R.; Rubin, B.P.; Mann, G.N. Epithelioid Sarcoma: The University of Washington Experience. Am. J. Surg. 2008, 196, 407–412.
- Guzzetta, A.A.; Montgomery, E.A.; Lyu, H.; Hooker, C.M.; Meyer, C.F.; Loeb, D.M.; Frassica, D.; Weber, K.L.; Ahuja, N. Epithelioid Sarcoma: One Institution’s Experience with a Rare Sarcoma. J. Surg. Res. 2012, 177, 116–122.
- Levy, A.; Le Péchoux, C.; Terrier, P.; Bouaita, R.; Domont, J.; Mir, O.; Coppola, S.; Honoré, C.; Le Cesne, A.; Bonvalot, S. Epithelioid Sarcoma: Need for a Multimodal Approach to Maximize the Chances of Curative Conservative Treatment. Ann. Surg. Oncol. 2014, 21, 269–276.
- Pradhan, A.; Grimer, R.J.; Abudu, A.; Tillman, R.M.; Carter, S.R.; Jeys, L.; Ferguson, P.C.; Griffin, A.M.; Wunder, J.S. Epithelioid Sarcomas: How Important Is Loco-Regional Control? Eur. J. Surg. Oncol. 2017, 43, 1746–1752.
- Elsamna, S.T.; Amer, K.; Elkattawy, O.; Beebe, K.S. Epithelioid Sarcoma: Half a Century Later. Acta Oncol. 2020, 59, 48–54.
- Frezza, A.M.; Sbaraglia, M.; Lo Vullo, S.; Baldi, G.G.; Simeone, N.; Frenos, F.; Campanacci, D.; Stacchiotti, S.; Pasquali, S.; Callegaro, D.; et al. The Natural History of Epithelioid Sarcoma. A Retrospective Multicentre Case-Series within the Italian Sarcoma Group. Eur. J. Surg. Oncol. 2020, 46, 1320–1326.
- Medjkane, S.; Novikov, E.; Versteege, I.; Delattre, O. The Tumor Suppressor HSNF5/INI1 Modulates Cell Growth and Actin Cytoskeleton Organization. Cancer Res. 2004, 64, 3406–3413.
- Vries, R.G.J. Cancer-Associated Mutations in Chromatin Remodeler HSNF5 Promote Chromosomal Instability by Compromising the Mitotic Checkpoint. Genes Dev. 2005, 19, 665–670.
- Wang, X.; Haswell, J.R.; Roberts, C.W.M. Molecular Pathways: SWI/SNF (BAF) Complexes Are Frequently Mutated in Cancer-Mechanisms and Potential Therapeutic Insights. Clin. Cancer Res. 2014, 20, 21–27.
- Raab, J.R.; Resnick, S.; Magnuson, T. Genome-Wide Transcriptional Regulation Mediated by Biochemically Distinct SWI/SNF Complexes. PLoS Genet. 2015, 11, e1005748.
- Alpsoy, A.; Dykhuizen, E.C. Glioma Tumor Suppressor Candidate Region Gene 1 (GLTSCR1) and Its Paralog GLTSCR1-like Form SWI/SNF Chromatin Remodeling Subcomplexes. J. Biol. Chem. 2018, 293, 3892–3903.
- Michel, B.C.; D’Avino, A.R.; Cassel, S.H.; Mashtalir, N.; McKenzie, Z.M.; McBride, M.J.; Valencia, A.M.; Zhou, Q.; Bocker, M.; Soares, L.M.M.; et al. A Non-Canonical SWI/SNF Complex Is a Synthetic Lethal Target in Cancers Driven by BAF Complex Perturbation. Nat. Cell Biol. 2018, 20, 1410–1420.
- Centore, R.C.; Sandoval, G.J.; Soares, L.M.M.; Kadoch, C.; Chan, H.M. Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet. TIG 2020, 36, 936–950.
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and Bioinformatic Analysis of Mammalian SWI/SNF Complexes Identifies Extensive Roles in Human Malignancy. Nat. Genet. 2013, 45, 592–601.
- Thway, K.; Jones, R.L.; Noujaim, J.; Fisher, C. Epithelioid Sarcoma: Diagnostic Features and Genetics. Adv. Anat. Pathol. 2016, 23, 41–49.
- Hollmann, T.J.; Hornick, J.L. INI1-Deficient Tumors: Diagnostic Features and Molecular Genetics. Am. J. Surg. Pathol. 2011, 35, e47–e63.
- Agaimy, A. The Expanding Family of SMARCB1(INI1)-Deficient Neoplasia: Implications of Phenotypic, Biological, and Molecular Heterogeneity. Adv. Anat. Pathol. 2014, 21, 394–410.
- Margol, A.S.; Judkins, A.R. Pathology and Diagnosis of SMARCB1-Deficient Tumors. Cancer Genet. 2014, 207, 358–364.
- Sullivan, L.M.; Folpe, A.L.; Pawel, B.R.; Judkins, A.R.; Biegel, J.A. Epithelioid Sarcoma Is Associated with a High Percentage of SMARCB1 Deletions. Mod. Pathol. 2013, 26, 385–392.
- Le Loarer, F.; Zhang, L.; Fletcher, C.D.; Ribeiro, A.; Singer, S.; Italiano, A.; Neuville, A.; Houlier, A.; Chibon, F.; Coindre, J.-M.; et al. Consistent SMARCB1 Homozygous Deletions in Epithelioid Sarcoma and in a Subset of Myoepithelial Carcinomas Can Be Reliably Detected by FISH in Archival Material. Genes Chromosomes Cancer 2014, 53, 475–486.
- Gounder, M.; Schöffski, P.; Jones, R.L.; Agulnik, M.; Cote, G.M.; Villalobos, V.M.; Attia, S.; Chugh, R.; Chen, T.W.-W.; Jahan, T.; et al. Tazemetostat in Advanced Epithelioid Sarcoma with Loss of INI1/SMARCB1: An International, Open-Label, Phase 2 Basket Study. Lancet Oncol. 2020, 21, 1423–1432.
- Papp, G.; Changchien, Y.-C.; Péterfia, B.; Pecsenka, L.; Krausz, T.; Stricker, T.P.; Khoor, A.; Donner, L.; Sápi, Z. SMARCB1 Protein and MRNA Loss Is Not Caused by Promoter and Histone Hypermethylation in Epithelioid Sarcoma. Mod. Pathol. 2013, 26, 393–403.
- Jamshidi, F.; Bashashati, A.; Shumansky, K.; Dickson, B.; Gokgoz, N.; Wunder, J.S.; Andrulis, I.L.; Lazar, A.J.; Shah, S.P.; Huntsman, D.G.; et al. The Genomic Landscape of Epithelioid Sarcoma Cell Lines and Tumours. J. Pathol. 2016, 238, 63–73.
- Kohashi, K.; Yamamoto, H.; Kumagai, R.; Yamada, Y.; Hotokebuchi, Y.; Taguchi, T.; Iwamoto, Y.; Oda, Y. Differential MicroRNA Expression Profiles between Malignant Rhabdoid Tumor and Epithelioid Sarcoma: MiR193a-5p Is Suggested to Downregulate SMARCB1 MRNA Expression. Mod. Pathol. 2014, 27, 832–839.
- Papp, G.; Krausz, T.; Stricker, T.P.; Szendrői, M.; Sápi, Z. SMARCB1 Expression in Epithelioid Sarcoma Is Regulated by MiR-206, MiR-381, and MiR-671-5p on Both MRNA and Protein Levels: Smarcb1 Regulation By Mirnas In Epithelioid Sarcoma. Genes Chromosomes Cancer 2014, 53, 168–176.
- Sápi, Z.; Papp, G.; Szendrői, M.; Pápai, Z.; Plótár, V.; Krausz, T.; Fletcher, C.D.M. Epigenetic Regulation of SMARCB1 By MiR-206, -381 and -671-5p Is Evident in a Variety of SMARCB1 Immunonegative Soft Tissue Sarcomas, While MiR-765 Appears Specific for Epithelioid Sarcoma. A MiRNA Study of 223 Soft Tissue Sarcomas. Genes Chromosomes Cancer 2016, 55, 786–802.
- Brenca, M.; Rossi, S.; Lorenzetto, E.; Piccinin, E.; Piccinin, S.; Rossi, F.M.; Giuliano, A.; Dei Tos, A.P.; Maestro, R.; Modena, P. SMARCB1/INI1 Genetic Inactivation Is Responsible for Tumorigenic Properties of Epithelioid Sarcoma Cell Line VAESBJ. Mol. Cancer Ther. 2013, 12, 1060–1072.
- Cooper, G.W.; Hong, A.L. SMARCB1-Deficient Cancers: Novel Molecular Insights and Therapeutic Vulnerabilities. Cancers 2022, 14, 3645.
- Versteege, I.; Medjkane, S.; Rouillard, D.; Delattre, O. A Key Role of the HSNF5/INI1 Tumour Suppressor in the Control of the G1-S Transition of the Cell Cycle. Oncogene 2002, 21, 6403–6412.
- Lee, S.; Cimica, V.; Ramachandra, N.; Zagzag, D.; Kalpana, G.V. Aurora A Is a Repressed Effector Target of the Chromatin Remodeling Protein INI1/HSNF5 Required for Rhabdoid Tumor Cell Survival. Cancer Res. 2011, 71, 3225–3235.
- Lin, L.; Hicks, D.; Xu, B.; Sigel, J.E.; Bergfeld, W.F.; Montgomery, E.; Fisher, C.; Hartke, M.; Tubbs, R.; Goldblum, J.R. Expression Profile and Molecular Genetic Regulation of Cyclin D1 Expression in Epithelioid Sarcoma. Mod. Pathol. 2005, 18, 705–709.
- Isakoff, M.S.; Sansam, C.G.; Tamayo, P.; Subramanian, A.; Evans, J.A.; Fillmore, C.M.; Wang, X.; Biegel, J.A.; Pomeroy, S.L.; Mesirov, J.P.; et al. Inactivation of the Snf5 Tumor Suppressor Stimulates Cell Cycle Progression and Cooperates with P53 Loss in Oncogenic Transformation. Proc. Natl. Acad. Sci. USA 2005, 102, 17745–17750.
- Stojanova, A.; Tu, W.B.; Ponzielli, R.; Kotlyar, M.; Chan, P.-K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Raught, B.; Penn, L.Z. MYC Interaction with the Tumor Suppressive SWI/SNF Complex Member INI1 Regulates Transcription and Cellular Transformation. Cell Cycle 2016, 15, 1693–1705.
- Weissmiller, A.M.; Wang, J.; Lorey, S.L.; Howard, G.C.; Martinez, E.; Liu, Q.; Tansey, W.P. Inhibition of MYC by the SMARCB1 Tumor Suppressor. Nat. Commun. 2019, 10, 2014.
- Msaouel, P.; Malouf, G.G.; Su, X.; Yao, H.; Tripathi, D.N.; Soeung, M.; Gao, J.; Rao, P.; Coarfa, C.; Creighton, C.J.; et al. Comprehensive Molecular Characterization Identifies Distinct Genomic and Immune Hallmarks of Renal Medullary Carcinoma. Cancer Cell 2020, 37, 720–734.e13.
- Lee, D.; Kim, J.W.; Seo, T.; Hwang, S.G.; Choi, E.-J.; Choe, J. SWI/SNF Complex Interacts with Tumor Suppressor P53 and Is Necessary for the Activation of P53-Mediated Transcription. J. Biol. Chem. 2002, 277, 22330–22337.
- Ray, A.; Mir, S.N.; Wani, G.; Zhao, Q.; Battu, A.; Zhu, Q.; Wang, Q.-E.; Wani, A.A. Human SNF5/INI1, a Component of the Human SWI/SNF Chromatin Remodeling Complex, Promotes Nucleotide Excision Repair by Influencing ATM Recruitment and Downstream H2AX Phosphorylation. Mol. Cell. Biol. 2009, 29, 6206–6219.
- Fontana, G.A.; Rigamonti, A.; Lenzken, S.C.; Filosa, G.; Alvarez, R.; Calogero, R.; Bianchi, M.E.; Barabino, S.M.L. Oxidative Stress Controls the Choice of Alternative Last Exons via a Brahma-BRCA1-CstF Pathway. Nucleic Acids Res. 2017, 45, 902–914.
- Jagani, Z.; Mora-Blanco, E.L.; Sansam, C.G.; McKenna, E.S.; Wilson, B.; Chen, D.; Klekota, J.; Tamayo, P.; Nguyen, P.T.L.; Tolstorukov, M.; et al. Loss of the Tumor Suppressor Snf5 Leads to Aberrant Activation of the Hedgehog-Gli Pathway. Nat. Med. 2010, 16, 1429–1433.
- Mora-Blanco, E.L.; Mishina, Y.; Tillman, E.J.; Cho, Y.-J.; Thom, C.S.; Pomeroy, S.L.; Shao, W.; Roberts, C.W.M. Activation of β-Catenin/TCF Targets Following Loss of the Tumor Suppressor SNF5. Oncogene 2014, 33, 933–938.
- Choi, S.K.; Kim, M.J.; You, J.S. SMARCB1 Acts as a Quiescent Gatekeeper for Cell Cycle and Immune Response in Human Cells. Int. J. Mol. Sci. 2020, 21, 3969.
- Alimova, I.; Birks, D.K.; Harris, P.S.; Knipstein, J.A.; Venkataraman, S.; Marquez, V.E.; Foreman, N.K.; Vibhakar, R. Inhibition of EZH2 Suppresses Self-Renewal and Induces Radiation Sensitivity in Atypical Rhabdoid Teratoid Tumor Cells. Neuro-Oncology 2013, 15, 149–160.
- Wilson, B.G.; Wang, X.; Shen, X.; McKenna, E.S.; Lemieux, M.E.; Cho, Y.-J.; Koellhoffer, E.C.; Pomeroy, S.L.; Orkin, S.H.; Roberts, C.W.M. Epigenetic Antagonism between Polycomb and SWI/SNF Complexes during Oncogenic Transformation. Cancer Cell 2010, 18, 316–328.
- Joldoshova, A.; Elzamly, S.; Brown, R.; Buryanek, J. Prometastatic CXCR4 and Histone Methyltransferase EZH2 Are Upregulated in SMARCB1/INI1-Deficient and TP53-Mutated Poorly Differentiated Chordoma. J. Mol. Pathol. 2022, 3, 7.
- Kadoch, C.; Copeland, R.A.; Keilhack, H. PRC2 and SWI/SNF Chromatin Remodeling Complexes in Health and Disease. Biochemistry 2016, 55, 1600–1614.
- Kadoch, C.; Crabtree, G.R. Mammalian SWI/SNF Chromatin Remodeling Complexes and Cancer: Mechanistic Insights Gained from Human Genomics. Sci. Adv. 2015, 1, e1500447.
- Völkel, P.; Dupret, B.; Le Bourhis, X.; Angrand, P.-O. Diverse Involvement of EZH2 in Cancer Epigenetics. Am. J. Transl. Res. 2015, 7, 175–193.
- Knutson, S.K.; Warholic, N.M.; Wigle, T.J.; Klaus, C.R.; Allain, C.J.; Raimondi, A.; Porter Scott, M.; Chesworth, R.; Moyer, M.P.; Copeland, R.A.; et al. Durable Tumor Regression in Genetically Altered Malignant Rhabdoid Tumors by Inhibition of Methyltransferase EZH2. Proc. Natl. Acad. Sci. USA 2013, 110, 7922–7927.
- Stacchiotti, S.; Zuco, V.; Tortoreto, M.; Cominetti, D.; Frezza, A.M.; Percio, S.; Indio, V.; Barisella, M.; Monti, V.; Brich, S.; et al. Comparative Assessment of Antitumor Effects and Autophagy Induction as a Resistance Mechanism by Cytotoxics and EZH2 Inhibition in INI1-Negative Epithelioid Sarcoma Patient-Derived Xenograft. Cancers 2019, 11, 1015.
- Italiano, A.; Soria, J.-C.; Toulmonde, M.; Michot, J.-M.; Lucchesi, C.; Varga, A.; Coindre, J.-M.; Blakemore, S.J.; Clawson, A.; Suttle, B.; et al. Tazemetostat, an EZH2 Inhibitor, in Relapsed or Refractory B-Cell Non-Hodgkin Lymphoma and Advanced Solid Tumours: A First-in-Human, Open-Label, Phase 1 Study. Lancet Oncol. 2018, 19, 649–659.





