Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Prodromos Hytiroglou | -- | 1842 | 2022-08-26 13:21:26 | | | |
| 2 | Catherine Yang | Meta information modification | 1842 | 2022-08-29 03:39:00 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Hytiroglou, P.; Bioulac-Sage, P.; Theise, N.D.; Sempoux, C. Hepatocellular Neoplasms. Encyclopedia. Available online: https://encyclopedia.pub/entry/26548 (accessed on 08 February 2026).
Hytiroglou P, Bioulac-Sage P, Theise ND, Sempoux C. Hepatocellular Neoplasms. Encyclopedia. Available at: https://encyclopedia.pub/entry/26548. Accessed February 08, 2026.
Hytiroglou, Prodromos, Paulette Bioulac-Sage, Neil D. Theise, Christine Sempoux. "Hepatocellular Neoplasms" Encyclopedia, https://encyclopedia.pub/entry/26548 (accessed February 08, 2026).
Hytiroglou, P., Bioulac-Sage, P., Theise, N.D., & Sempoux, C. (2022, August 26). Hepatocellular Neoplasms. In Encyclopedia. https://encyclopedia.pub/entry/26548
Hytiroglou, Prodromos, et al. "Hepatocellular Neoplasms." Encyclopedia. Web. 26 August, 2022.
Copy Citation
Hepatocellular carcinoma (HCC), a major global contributor of cancer death, usually arises in a background of chronic liver disease, as a result of molecular changes that deregulate important signal transduction pathways. Certain molecular changes of hepatocarcinogenesis are associated with clinicopathologic features and prognosis, suggesting that subclassification of HCC is practically useful. On the other hand, subclassification of hepatocellular adenomas (HCAs), a heterogenous group of neoplasms, has been well established on the basis of genotype–phenotype correlations.
hepatocellular adenoma
hepatocellular carcinoma
molecular pathology
etiology
pathogenesis
diagnosis
1. Etiology and Pathogenesis of Hepatocellular Adenomas
It is now well recognized that hepatocellular adenoma (HCA), occurring mainly in young women taking oral contraception (OC), is a heterogeneous entity comprising different morpho-molecular subtypes, with various clinical and etiological backgrounds, risk for complications (bleeding and malignant transformation), and pathogenesis [1]. While most HCAs appear in normal liver, several clinical conditions and genetic syndromes have also been found to be linked to the development of HCAs [1].
The first well-recognized subtype is related to HNF1A-inactivating mutations (H-HCA). These tumors may be solitary or multiple, or they may occur in the context of liver adenomatosis. H-HCA is usually characterized by steatosis within the lesion and has a low risk of complications.
The second subtype is the inflammatory hepatocellular adenoma (IHCA), often developing on a background of NAFLD or in the context of alcohol consumption, predominantly but not exclusively in obese women. These lesions are often multiple. Typically characterized by sinusoidal dilatation and inflammation, IHCAs are related to different mutations leading to IL6/JAK/STAT inflammatory pathway activation.
A third subtype is the HCA with β-catenin-activating mutations (b-HCA). A proportion of these mutations occur in IHCA, thus giving rise to b-IHCA. By contrast with the other subtypes, b-(I)HCAs are overrepresented in men and have a higher risk of malignant transformation. This risk depends on the level of activation of the β-catenin pathway, which is linked to the type of CTNNB1 mutation that results also in different immunohistochemical features [2].
A recently identified fourth HCA subtype is related to activation of the sonic hedgehog pathway (shHCA). These tumors are prone to bleeding, even when small, and can be recognized by argininosuccinate synthase 1 (ASS1) overexpression on immunohistochemistry [3][4]. This subtype has been described so far only in women, often overweight, and in the context of the metabolic syndrome.
Figure 1 illustrates the different subtypes of HCA with their principal immunohistochemical characteristics: H-HCA and liver fatty-acid-binding protein (LFABP), IHCA and C-reactive protein (CRP), b-HCA/b-IHCA and glutamine synthetase (GS), and shHCA and ASS1.
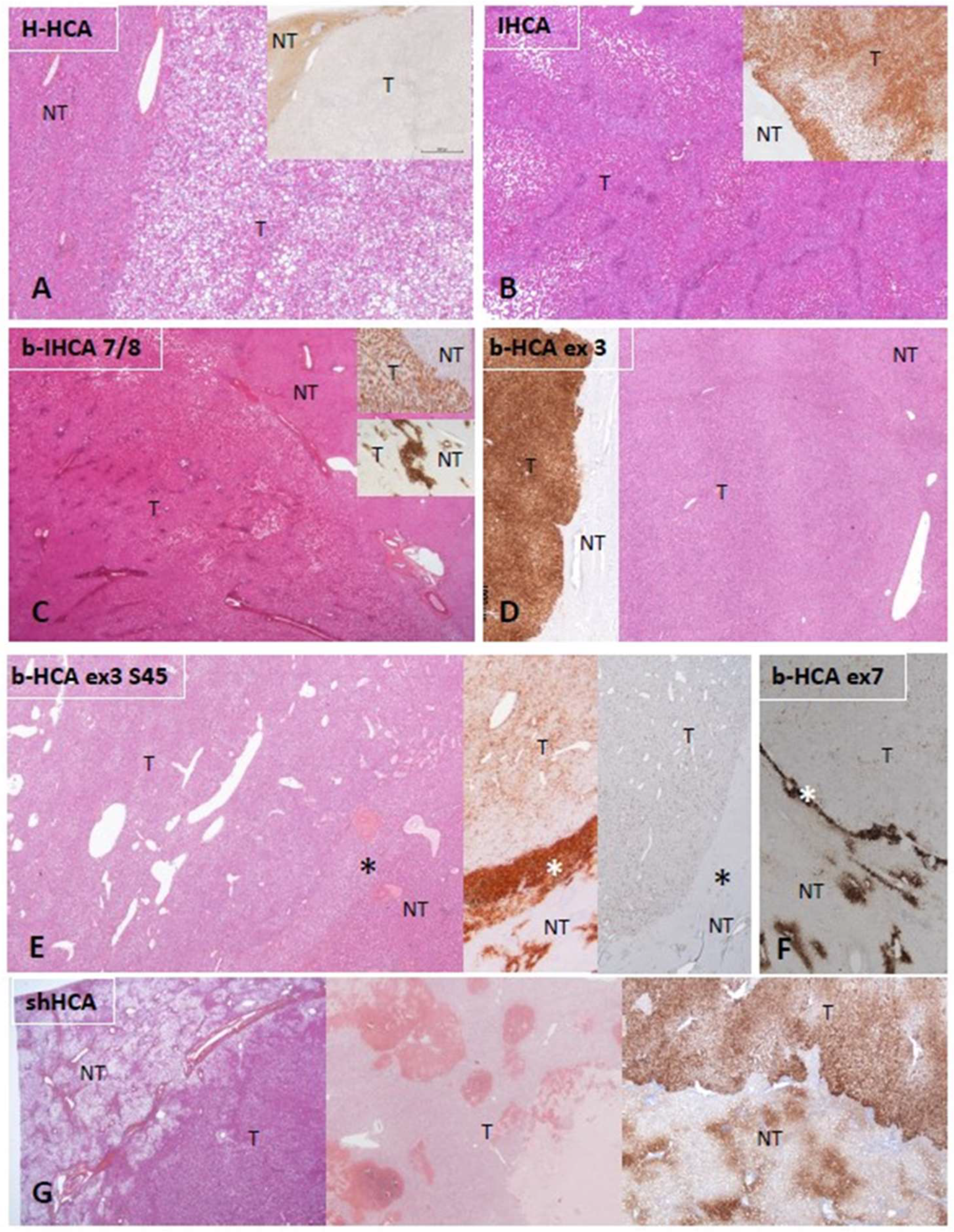
Figure 1. Characteristic histologic and immunohistochemical features of HCAs. (A) H-HCA: The tumor (T) appears highly steatotic on H&E with a complete lack of LFABP by immunohistochemistry (insert), contrasting with the normal expression in the nontumorous liver (NT). (B) IHCA: The tumor (T) exhibits sinusoidal dilatation on H&E with a strong CRP expression by immunohistochemistry (insert), sharply demarcated from the nontumorous liver (NT). (C) CTNNB1 exon 7/8 mutated b-IHCA: This tumor exhibits a classical appearance of IHCA (sinusoidal dilatation, numerous thick arteries, and strong expression of CRP (top insert); in addition, GS is very faint in the tumor but with a strong GS rim between tumor (T) and nontumorous liver (NT) (bottom insert); molecular analysis identified a mutation on CTNNB1 exon 7/8 (see [2]). (D) CTNNB1 exon 3 mutated b-HCA: This tumor (T), which is not well delimited from the nontumorous liver (NT) on H&E, exhibits a strong and diffuse GS expression (left insert), identifying a high level of activation of the β-catenin pathway (large deletion on exon 3). (E) Exon 3 S45 mutated b-HCA: This tumor (T) exhibits numerous irregular vessels below the rim (asterisk) that separates T from nontumorous liver (NT); heterogeneous expression of GS is seen in T, whereas a strong GS expression characterizes the rim (middle insert); a corresponding diffuse CD34 immunostaining is seen in the endothelial cells of T, with no CD34 expression in the rim (asterisk) (right insert) (see [2]). (F) Exon 7 mutated b-HCA: GS is very faint in the tumor (T), and a thin GS rim (asterisk) separates T from the nontumorous liver (NT); molecular methods identified a β-catenin exon 7 mutation. (G) shHCA: This tumor developed in a highly steatotic nontumorous liver (NT, left picture) and exhibits focally large hemorrhagic foci (middle picture); ASS1 immunohistochemistry shows an overexpression in the tumor (T), in comparison with the nontumorous liver (NT), in which its expression is restricted to the periportal/septal zones (right picture). Abbreviations: H-HCA, HNF1A-mutated hepatocellular adenoma; IHCA, inflammatory HCA; b-IHCA, β-catenin-mutated inflammatory HCA; b-HCA, β-catenin-mutated HCA; shHCA, sonic hedgehog-activated HCA; LFABP, liver fatty-acid-binding protein; CRP, C reactive protein; GS, glutamine synthetase.
2. Diagnosis and Subtyping of Hepatocellular Adenomas
The histologic diagnosis of HCA requires careful assessment of representative hematoxylin and eosin (H&E)-stained sections. HCAs are characterized by a benign hepatocellular proliferation, devoid of portal tracts. “Unpaired” arteries (i.e., arteries unaccompanied by veins or bile ducts) are present among the neoplastic cells. Other characteristic features include steatosis, inflammation, sinusoidal dilatation, and/or areas of hemorrhage. After H&E assessment, immunohistochemical evaluation follows with specific antibodies recognizing the targets identified by the genotype–phenotype studies [1][2]. An algorithm for the diagnosis is proposed in Figure 2.
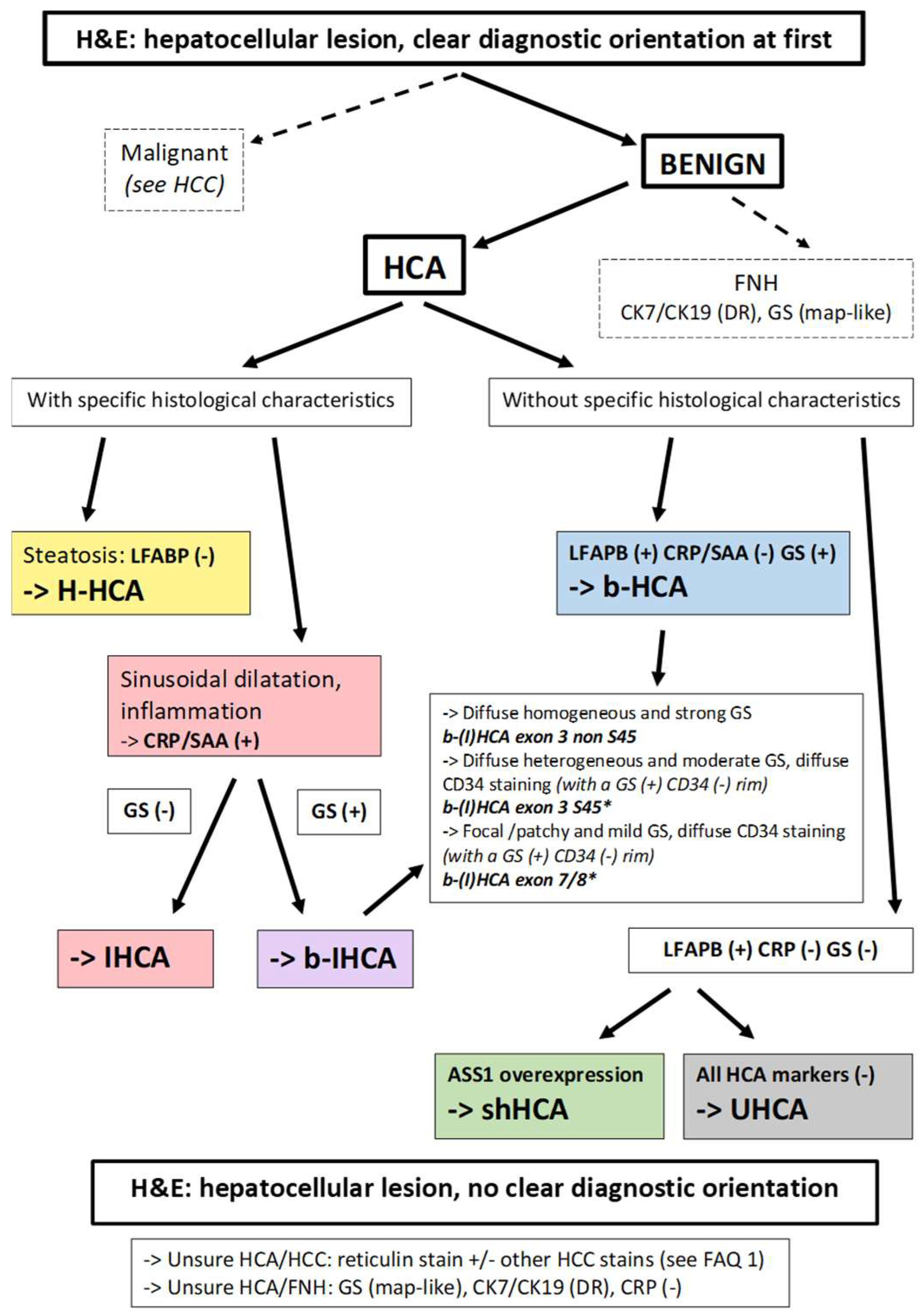
Figure 2. Diagnostic algorithm for HCAs. From a practical point of view, most of the cases are easily recognized as benign or malignant, but some are not. In the situation of an obvious HCA, if there is steatosis, with LFABP (−) and GS (−), there is no need to perform further IHC staining; it can be concluded that the tumor is an H-HCA. If an HCA shows sinusoidal dilatation and inflammation, with LFABP (+) and GS (−), it is mandatory to perform CRP and/or SAA immunostaining in order to diagnose an IHCA. GS immunostaining is mandatory in all IHCAs in order to diagnose a b-IHCA. Different patterns of GS staining exist, linked to the type of underlying mutations (see [2]). If LFABP is positive and all other markers are negative, then an overexpression of ASS1 will lead to the identification of a shHCA, whereas, if it is not overexpressed, it is an UHCA. * Importantly, the GS(+)/CD34(−) rim can be irregular or discontinuous and is usually better represented in b-HCA than in b-IHCA. Its recognition on biopsies can be challenging (see [2]). In case of an uncertain diagnosis, HCA versus HCC or HCA versus FNH, additional histochemical and immunohistochemical stains are needed. The differential diagnosis of HCA versus HCC is discussed in FAQ 1. Reticulin stain might help to recognize alterations of the framework, although it is not a strict feature. Cytokeratin 7 and cytokeratin 19 stains help to recognize ductular reaction, and GS has a specific map-like pattern in FNH. Abbreviations: HCA, hepatocellular adenoma; H-HCA, HNF1A-mutated HCA; IHCA, inflammatory HCA; b-HCA, β-catenin-activated HCA; b-IHCA, β-catenin-activated and inflammatory HCA; shHCA, sonic hedgehog-activated HCA; UHCA, unclassified HCA; HCC, hepatocellular carcinoma; FNH, focal nodular hyperplasia; LFABP, liver fatty-acid-binding protein; CRP, C reactive protein; SAA, serum amyloid A; GS, glutamine synthetase; ASS1, argininosuccinate synthase; CK7, cytokeratin 7; CK19, cytokeratin 19; DR, ductular reaction.
3. Etiology and Pathogenesis of Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) usually arises in livers with chronic disease, and it is most often discovered when disease has reached an advanced stage, traditionally known as cirrhosis. The most common chronic diseases that are associated with HCC are chronic hepatitis B, chronic hepatitis C, and alcoholic liver disease, accounting together for 84% of the cases occurring globally in 2015 [5]. In the meanwhile, nonalcoholic steatohepatitis (NASH), associated with the metabolic syndrome, is emerging as a major risk factor for HCC [6]. Other risk factors include hereditary metabolic disorders (such as hemochromatosis, α1-antitrypsin deficiency, and tyrosinemia), aflatoxin B1 exposure (in individuals chronically infected with HBV), and tobacco smoking. Chronic liver diseases other than those mentioned above (e.g., autoimmune hepatitis, primary biliary cholangitis, primary sclerosing cholangitis, and Wilson disease) are uncommonly associated with development of HCC.
HCC arising in noncirrhotic livers is often caused by HBV, which is a virus with known carcinogenic effects. HBV DNA insertion in the host genome can deregulate genes involved in cell signaling and replication (such as TERT, PDGFR, MLL4, and CCNE1), while the HBV X protein transactivates genes involved in signal transduction pathways and inhibits TP53 expression [7][8][9]. NASH and hereditary hemochromatosis are also increasingly recognized as causes of HCC arising in noncirrhotic livers [6][10]. However, HCC can also arise in apparently normal liver. Some of these cases may represent evolution of HCA (mostly b-HCA and b-IHCA) to HCC (discussed in the previous sections), while others, usually occurring in older individuals, remain unexplained. A special HCC subtype arising in normal livers of young individuals is fibrolamellar carcinoma, which is associated with a characteristic somatic gene fusion, DNAJB1–PRKACA, resulting from deletions in chromosome 19 and activating protein kinase A [11].
In chronic liver diseases, continuous cell loss results in cell proliferation occurring in a noxious microenvironment, characterized by oxidative stress due to chronic inflammation, overexpression of growth factors, and epigenetic changes due to derangements of DNA methyltransferases [12][13][14]. Thus, the possibility of mutations that initiate or promote carcinogenesis is increased, while mutations providing survival benefits to hepatocytes favor clonal expansion. This process is accelerated in the advanced stages of chronic liver diseases when vascular changes, including intrahepatic vein thrombosis and vascular reorganization, result in extensive cell loss. In that setting, hepatic regeneration largely depends on progenitor cell proliferation due to senescence of hepatocytes. Therefore, critical mutations in progenitor cells have the potential to produce large numbers of clonally expanding hepatocytes with increased likelihood to progress to precancerous lesions and then to HCC.
The diverse molecular changes that are associated with HCC have been recently reviewed [15]. Whole-exome and whole-genome sequencing studies have revealed 40–60 somatic coding mutations per HCC, including 4–6 driver mutations [16]. The most frequent mutations in HCC are those involving the promoter of telomerase reverse transcriptase (TERT), occurring in 60% of cases [17]. In an additional 30% of HCCs, TERT is deregulated by other molecular mechanisms, such as viral insertion [18]. TERT promoter mutations have also been detected in precancerous nodules and are considered an early event in hepatocarcinogenesis [19]. Other frequently mutated genes in HCC include CTNNB1, TP53, RB1, ARID1A, ARID2, AXIN1, albumin, and apolipoprotein B [20][21][22]. The mutations occurring in hepatocarcinogenesis can disrupt various signal transduction pathways, such as telomere maintenance (TERT), cell-cycle control (TP53, CDKN2A), Wnt/β-catenin (CTNNB1, AXIN1), epigenetic (ARID1A, ARID2, MLL2), and oxidative stress (NFE2L2, KEAP1) [17][23][24]. “Druggable” genetic alterations are under intense investigation because, at the present time, targeted therapeutic agents for HCC are limited to a small number of multikinase inhibitors. On the other hand, understanding the interaction between neoplastic cells and their microenvironment will be crucial for identifying biomarkers and developing new therapies based on immune checkpoint inhibition [25]
Recent studies have shown that certain molecular changes in HCC are associated with specific clinicopathologic features and prognosis, suggesting the possibility of a molecular classification for the future [26][27][28][29]. This active research has resulted in the recognition of several HCC subtypes (also called “variants”) that hold promise for a more personalized treatment of HCC patients. Eight HCC subtypes, considered to represent distinct clinicopathological/molecular entities and accounting together for up to 35% of HCCs, have been included in the latest edition of the WHO classification of liver tumors [30].
References
- Bioulac-Sage, P.; Gouw, A.S.; Balabaud, C.; Sempoux, C. Hepatocellular adenoma: What we know, what we do not know, and why it matters. Histopathology 2022, 80, 878–897.
- Sempoux, C.; Gouw, A.S.; Dunet, V.; Paradis, V.; Balabaud, C.; Bioulac-Sage, P. Predictive Patterns of Glutamine Synthetase Immunohistochemical Staining in CTNNB1-mutated Hepatocellular Adenomas. Am. J. Surg. Pathol. 2021, 45, 477–487.
- Henriet, E.; Hammoud, A.A.; Dupuy, J.-W.; Dartigues, B.; Ezzoukry, Z.; Dugot-Senant, N.; Leste-Lasserre, T.; Pallares-Lupon, N.; Nikolski, M.; Le Bail, B.; et al. Argininosuccinate synthase 1 (ASS1): A marker of unclassified hepatocellular adenoma and high bleeding risk. Hepatology 2017, 66, 2016–2028.
- Sala, M.; Gonzales, D.; Leste-Lasserre, T.; Dugot-Senant, N.; Paradis, V.; Di Tommaso, S.; Dupuy, J.; Pitard, V.; Dourthe, C.; Sciarra, A.; et al. ASS1 Overexpression: A Hallmark of Sonic Hedgehog Hepatocellular Adenomas; Recommendations for Clinical Practice. Hepatol. Commun. 2020, 4, 809–824.
- Global Burden of Disease Liver Cancer Collaboration. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691.
- Ioannou, G.N. Epidemiology and risk-stratification of NAFLD-associated HCC. J. Hepatol. 2021, 75, 1476–1484.
- Paterlini-Bréchot, P.; Saigo, K.; Murakami, Y.; Chami, M.; Gozuacik, D.; Mugnier, C.; Lagorce, D.; Bréchot, C. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 2003, 22, 3911–3916.
- Murakami, Y.; Saigo, K.; Takashima, H.; Minami, M.; Okanoue, T.; Bréchot, C.; Brechot, P.P. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 2005, 54, 1162–1168.
- Sung, W.-K.; Zheng, H.; Li, S.; Chen, R.; Liu, X.; Li, Y.; Lee, N.P.; Lee, W.H.; Ariyaratne, P.N.; Tennakoon, C.; et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012, 44, 765–769.
- Goh, J.; Callagy, G.; McEntee, G.; O’Keane, J.C.; Bomford, A.; Crowe, J. Hepatocellular carcinoma arising in the absence of cirrhosis in genetic haemochromatosis: Three case reports and review of literature. Eur. J. Gastroenterol. Hepatol. 1999, 11, 915–919.
- Honeyman, J.N.; Simon, E.P.; Robine, N.; Chiaroni-Clarke, R.; Darcy, D.G.; Lim, I.I.P.; Gleason, C.E.; Murphy, J.M.; Rosenberg, B.R.; Teegan, L.; et al. Detection of a Recurrent DNAJB1-PRKACA Chimeric Transcript in Fibrolamellar Hepatocellular Carcinoma. Science 2014, 343, 1010–1014.
- Thorgeirsson, S.S.; Grisham, J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002, 31, 339–346.
- Hatziapostolou, M.; Polytarchou, C.; Aggelidou, E.; Drakaki, A.; Poultsides, G.A.; Jaeger, S.A.; Ogata, H.; Karin, M.; Struhl, K.; Hadzopoulou-Cladaras, M.; et al. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011, 147, 1233–1247.
- Brody, R.I.; Theise, N.D. An inflammatory proposal for hepatocarcinogenesis. Hepatology 2012, 56, 382–384.
- Hytiroglou, P.; Bioulac-Sage, P. Molecular Pathogenesis and Diagnostics of Hepatocellular Tumors. In Odze and Goldblum Surgical Pathology of the GI Tract, Liver, Biliary Tract, and Pancreas, 4th ed.; Odze, R.D., Goldblum, J.R., Eds.; Elsevier: Philadelphia, PA, USA, 2023; pp. 1369–1383.
- Gerbes, A.; Zoulim, F.; Tilg, H.; Dufour, J.-F.; Bruix, J.; Paradis, V.; Salem, R.; Peck-Radosavljevic, M.; Galle, P.R.; Greten, T.F.; et al. Gut roundtable meeting paper: Selected recent advances in hepatocellular carcinoma. Gut 2018, 67, 380–388.
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013, 4, 2218.
- Nault, J.-C.; Ningarhari, M.; Rebouissou, S.; Zucman-Rossi, J. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 544–558.
- Nault, J.C.; Calderaro, J.; Di Tommaso, L.; Balabaud, C.; Zafrani, E.S.; Bioulac-Sage, P.; Roncalli, M.; Zucman-Rossi, J. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 2014, 60, 1983–1992.
- The Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017, 169, 1327–1341.
- Dhanasekaran, R.; Nault, J.-C.; Roberts, L.R.; Zucman-Rossi, J. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology 2019, 156, 492–509.
- Nault, J.; Martin, Y.; Caruso, S.; Hirsch, T.; Bayard, Q.; Calderaro, J.; Charpy, C.; Copie-Bergman, C.; Ziol, M.; Bioulac-Sage, P.; et al. Clinical Impact of Genomic Diversity From Early to Advanced Hepatocellular Carcinoma. Hepatology 2020, 71, 164–182.
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H.; et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273.
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511.
- Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462.
- Boyault, S.; Rickman, D.S.; Bioulac-Sage, P.; De Reyniès, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007, 45, 42–52.
- Hoshida, Y.; Nijman, S.M.B.; Kobayashi, M.; Chan, J.A.; Brunet, J.-P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative Transcriptome Analysis Reveals Common Molecular Subclasses of Human Hepatocellular Carcinoma. Cancer Res. 2009, 69, 7385–7392.
- Nault, J.; De Reyniès, A.; Villanueva, A.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Decaens, T.; Franco, D.; Imbeaud, S.; Rousseau, F.; et al. A Hepatocellular Carcinoma 5-Gene Score Associated With Survival of Patients After Liver Resection. Gastroenterology 2013, 145, 176–187.
- Calderaro, J.; Couchy, G.; Imbeaud, S.; Amaddeo, G.; Letouzé, E.; Blanc, J.-F.; Laurent, C.; Hajji, Y.; Azoulay, D.; Bioulac-Sage, P.; et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 2017, 67, 727–738.
- Torbenson, M.S.; Ng, I.O.L.; Park, Y.N.; Roncalli, M.; Sakamoto, M. Hepatocellular carcinoma. In Digestive System Tumours, 5th ed.; WHO Classification of Tumours Editorial Board; WHO Classification of Tumours Series; International Agency for Research on Cancer: Lyon, France, 2019; Volume 1, pp. 229–239.
More
Information
Subjects:
Gastroenterology & Hepatology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Entry Collection:
Gastrointestinal Disease
Revisions:
2 times
(View History)
Update Date:
29 Aug 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




