
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Noboru Oriuchi | + 2849 word(s) | 2849 | 2020-10-07 04:41:31 | | | |
| 2 | Catherine Yang | Meta information modification | 2849 | 2020-10-22 04:35:17 | | |
Video Upload Options
Quantitative parameters of FDG-PET, such as SUV and TLG have been used to evaluate therapeutic response. Recent advancement in anti-cancer therapeutics showed that tumor response to molecular-targeted drugs and immune-checkpoint inhibitors is different from conventional chemotherapy in terms of temporal metabolic alteration and morphological change after the course of effective therapy. Metabolic changes and temporal enlargement due to immune cell infiltration seen after immune-checkpoint inhibitors, such as anti-programmed cell death-1 (PD-1) and anti-programmed cell death ligand 1 (PD-L1) antibodies facilitated the modification of conventional Response Evaluation Criteria in Solid Tumor and FDG-PET response evaluation criteria. Tumor microenvironment including cancer stem cells (CSCs) that is thought to be a root cause of tumor heterogeneity; is considered a target of novel and effective therapy.
Accumulation of FDG reflects glucose metabolism of both cancer cells and immunologically competent cells in the tumor microenvironment. Immunological reaction to the therapy differs among patients according to the individual immune function. Considering the heterogeneity of tumor tissue and individual variation in tumor response to immunotherapy, radiomics approach combines quantitative image features with deep learning algorithm have the potentials to improve response assessment in more personalized treatment.
Stromal cell-derived factor 1 (SDF-1)/C-X-C chemokine receptor type 4 (CXCR4)-targeted α-particle therapy has been introduced, because SDF-1/CXCR4 axis is known to increase epithelial-mesenchymal transition to facilitate invasion and metastasis, and regulate immune response by accelerating T cell proliferation as well as PD-1 and PD-L1 expression in cancer cells and cytotoxic T lymphocytes, respectively. Prominent energy profile and biological effect of α-particles are promising as an alternative in targeted radionuclide therapy (TRT). Radiation dosimetry according to the theranostics approach will permit accurate TRT and artificial intelligence-based treatment decision making and precise response evaluation would be a precision nuclear medicine in the future.
1. Introduction
Positron emission tomography (PET) has become an indispensable procedure for the initial assessment and post-therapeutic evaluation in clinical oncology, using dedicated radiopharmaceuticals targeting cellular metabolism and tumor-specific receptors [1]. PET as a means of molecular-based imaging is able to characterize biological processes associated with disease progression and therapeutic response quantitatively at the cellular and molecular levels. The outcome of a therapy cannot be interpreted properly without a surrogate biomarker to assess the efficacy of every therapeutic modality.
Therapeutic response is objectively evaluable by means of imaging. Conventional response evaluation criteria use morphological parameters; on the other hand, 2-[18F] fluoro-2-deoxy-d-glucose (FDG)-PET-based criteria use metabolic parameters. Histological response to anti-cancer therapy depends on the therapeutic modalities; cancer immunotherapy shows the distinctive phenomenon of immune-related tumor responses. Emerging observational data of immune-related response patterns have determined modification of the conventional response criteria. The current approaches to anti-cancer therapy target the tumor microenvironment as well as anti-tumor immunity.
2. Glucose Metabolism of Cancer and FDG-PET
It has been appreciated for nearly 100 years that cancer cells are metabolically distinct from other cells. All cells fundamentally require nutrients to meet metabolic demands for energy generation and biosynthesis. Metabolic demands of cell proliferation, differentiation, and biosynthesis of proteins, lipids, and nucleotides are different in tumor cells.
Elevated glucose uptake and cellular metabolism were thought to be the biochemical characteristics of cancer [2]. FDG-PET could disclose a high glycolytic rate and pyruvate oxidation in the mitochondria, depending on the cell proliferation. These altered metabolisms, including metabolic switch from aerobic to anaerobic glycolysis, are known as the Warburg effect [3][4]. The function of the Warburg effect has been simply understood as a metabolic switch, but a breakthrough to explain the Warburg effect regarding cancer metabolism in vivo has taken place recently [5][6][7][8].
Tumor hypoxia is known to be the most important factor to account for biological aggressiveness and resistance to chemotherapy and radiotherapy through the expression of multidrug resistance 1 (MDR1) and cell cycle arrest [8][9]. Accelerated proliferation and metabolism of cancer cells lead to an imbalance in the form of insufficient oxygen supply in relation to oxygen demand in solid tumors [10][11]. Anti-neoplastic drugs and ionizing radiation have effects on oxygen to generate reactive oxygen species (ROS) in cancer cells, causing oxidative stress, which results in apoptosis. However, cancer cells can survive in the hypoxic area, which is usually seen at 100 μm from tumor vessels, because of the decreased generation of ROS [12]. In the area of hypoxia, a transcription factor, hypoxia-inducible factor 1 (HIF-1), is activated to induce the expression of various genes responsible for adaption to hypoxic metabolism from oxidative phosphorylation to glycolytic ATP production, explained by the Warburg effect, as mentioned above [13][14], invasion and metastases of cancer cells through the formation of pre-metabolic niche and epithelial–mesenchymal transition (EMT) to escape from hypoxia [15][16], increased erythropoiesis through upregulation of erythropoietin, and angiogenesis to reoxygenation of hypoxic area [17]. An α-subunit of HIF-1 (HIF-1α) induces expression of glucose transporter 1 and glycolytic enzymes to increase glucose uptake and anaerobic glycolysis to compensate for ATP production [18][19]. FDG-PET can therefore evaluate tumor aggressiveness and resistance to chemotherapy and radiotherapy by detecting increased glucose metabolism and is a possible therapeutic marker to monitor responses.
3. Machine Learning for Imaging Cancer Heterogeneity and Interpretation
Figure 1 shows FDG-PET/CT images of a patient with NSCLC. Two foci of increased nodular FDG uptake are seen in the upper lobe of the right lung and the upper mediastinum on the left side. These lesions show SUVmax of 7.2 and 4.8, respectively. Do these images provide the radiologist with sufficient information for correct interpretation? Radiologists cannot diagnose correctly without additional information about clinical history, because every radiologist knows that the most common sites of metastasis of lung cancer are ipsilateral hilar and mediastinal lymph nodes and that metastasis to the contralateral mediastinal lymph node usually occurs after ipsilateral mediastinal lymph nodes metastases [20]. Information about past history of left lung cancer with T3N1M0, stage IIIA, for which left upper lobe segmentectomy and lymph node dissection was performed 2 years before, is a clue for correct diagnosis. The patient had undergone surgery followed by chemotherapy on the basis of correct diagnosis. How accurately can FDG-PET images estimate the efficacy of chemotherapy and prognosis of this patient? Do quantitative parameters help to predict response to chemotherapy and prognosis?
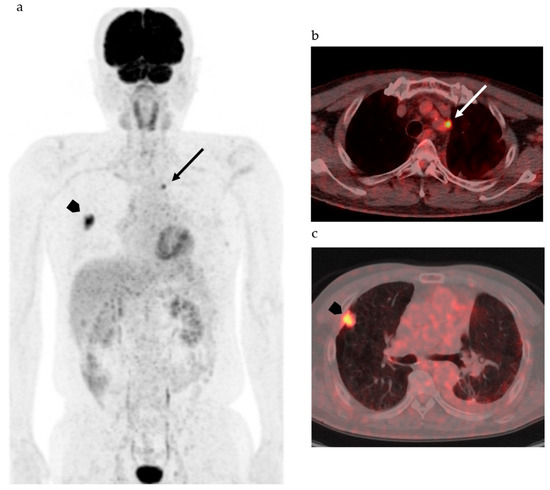
Figure 1. 2-[18F] fluoro-2-deoxy-d-glucose positron emission tomography/computed tomography (FDG-PET/CT) of patients with non-small cell lung cancer. (a) Maximum intense projection image shows abnormal FDG uptake in the left upper mediastinum (arrow) and the right lung (arrowhead). (b) PET/CT shows increased FDG uptake in the lymph node adjacent to the left subclavian artery at the level of the upper mediastinum (arrow). (c) High FDG uptake is seen in the nodule at the right upper lobe (arrow head).
Considering the heterogeneity of tumor tissue, more sophisticated indexes surpassing SUV and other parameters as well as diagnostic algorithms are needed to accurately classify the tumor on the basis of biological malignancy, effectiveness of various types of therapy, and prognosis of patients. Recent advancements in computer science and artificial intelligence (AI) have shown the possibility for machine learning systems to take on the practice of radiology, which was previously thought to be limited to human radiologists. AI including machine learning technologies has the potential to transform radiological imaging by using the vast amount of clinical data including pathologic and genetic examinations to automate the integrated diagnostic radiology workflow and diagnosis. Machine learning algorithms, such as random forests, support vector machines, and artificial neural networks, have been used for classification of images by training with input data set and knowledge, and then the best model is applied for the prediction of pathophysiology. Due to deep learning and convolutional neural networks (CNN), the capability to learn and master given tasks to perform computer-aided diagnosis (CAD) has made remarkable advances in clinical radiology in the past decade [21][22]. Wang et al. have suggested that the performance of CNN from FDG PET/CT images is comparable to the best classical machine learning and human radiologists and that CNN is more convenient and objective than the classical methods, because it does not need tumor segmentation, feature selection, or texture features for classifying mediastinal lymph node metastasis in patients with NSCLC [21]. They also suggested that the performance of the CNN would be improved by incorporating diagnostic features like SUV and tumor size [21]. For example, in mediastinal lymph node metastasis in patients with NSCLC, accurate diagnosis is a challenge, as indicated in Figure 1; however, lymph node metastasis evaluated by FDG uptake has been reported to be prognostic as compared with pathological lymph node metastasis [23]. Therefore, diagnosis of lymph node status during diagnostic work up is of the utmost importance.
Machine learning is already being applied in the practice of radiology, including in the field of mammography. There have been many papers describing a performance level in lesion detection similar to that of experienced radiologists [24][25][26]. CAD was approved by the Food and Drug Administration (FDA) and has been used for mammography in radiology practices [27]; however, improvement of the diagnostic ability has not been satisfactory, and the majority of radiologists have rarely changed their reports as a result of findings generated by CAD [28][29][30]. Machine learning has been reported to be unlikely to replace radiologists but will provide quantitative tools to increase the value of imaging as a biomarker including therapeutic response evaluation [31]. Recently, radiology professionals have reminded that AI algorithms must be as safe and effective as the physician by rigorous testing, longitudinal surveillance, and investigation of oversight mechanisms to ensure generalizability across patients as well as variable imaging and imaging protocols [32]. However, radiologists cannot disregard autonomous radiology AI, because AI can tirelessly improve the image reading capacity and may drastically acquire interpretation capabilities if AI can incorporate available medical information and contextual integration of data that would typically be identified during physician interpretation in order to render a medical judgement.
On the basis of considering tumor heterogeneity, texture analysis has been explored, especially in the field of nuclear medicine [32][33][34][35][36]. The most exciting part of machine learning in medical imaging would be to extract patterns that are beyond human perception and classification due to the application of deep learning for diagnostic algorithms [37][38]. Radiologists should seek to work alongside AI in the future.
4. Response Evaluation of Novel Therapeutics with Molecular Imaging
Malignant cells survive in a complex balance in the immune system. Both CTLA-4 and PD-1 suppress T cell activities. Therefore, agents that block CTLA-4, PD-1, and PD-L1 are able to produce an anti-tumor response through immune activation. Inhibition of CXCR4 exaggerates the anti-tumor immune response and CXCR4-targeted therapy is a possible therapeutic option to eradicate CSCs. Recent studies have indicated that dual blockade of PD-1–PD-L1 and CXCL-12–CXCR4 pathways reduces specific cellular and functional elements within the immunosuppressive tumor microenvironment and augments tumor-specific cell-mediated immune responses. The complexity of these interactions and heterogeneity of immune cells in the tumor microenvironment are challenges in the development and the evaluation of the therapeutic efficacy of new immune therapies in vivo. Imaging of immune cells that are major players in anti-cancer therapy is challenging because many subtypes of cells exist and play different roles in the tumor microenvironment.
Non-invasive evaluation procedures for therapy outcomes, such as biomarkers and molecular imaging, are expected to represent precise strategies of cancer therapy. FDG-PET can play an important role in fulfilling this purpose, as mentioned earlier [39][40][41]. Uptake of FDG reflects the viability of cancer cells and all other players of the immune system in the microenvironment. No uptake of FDG means complete remission of the tumor; however, increased uptake does not always indicate progression of the tumor, because of the pseudoprogression phenomenon and increased anaerobic glycolysis in the therapy-induced hypoxia, as mentioned above [18][39][42]. Cancer cell-specific imaging has the potential to evaluate quantitatively the residual cancer cells that had been able to evade anti-cancer agent of immune response. However, phenotypic changes due to genetic alteration, such as therapy resistant mutation and de novo mutation after therapy, may decrease specificity to the specific imaging agent. Metabolism-based PET tracers other than FDG can be used to evaluate therapeutic efficacy [43][44]. However, metabolic diversity and instability, especially those acquired on the progression course or after therapy, of cancer cells would be sources of inaccuracy in evaluating the response.
Prediction and evaluation of therapeutic efficacy would be possible with a tumor-specific PET tracer. Prostate-specific membrane antigen (PSMA) ligand labeled with gallium-68 (68Ga-PSMA) is a PET tracer used to determine the eligibility for PSMA-targeted radionuclide therapy with 177Lu-PSMA or 235Ac-PSMA (Table 1). Peptide receptor radionuclide therapy for neuroendocrine carcinoma with 90Y- and 177Lu-dodecane-tetraacetic acid-Tyr3-octreotate (DOTA-TATE) is another radionuclide therapy performed successfully for solid tumors. 68Ga-DOTA-TATE is a diagnostic counterpart of therapeutics. These examples are representative theranostics in nuclear medicine practice that will be followed by the future radionuclide therapy. A major role of specific imaging in the theranostics is to confirm the indication of therapy. Another role would be dosimetry analysis to determine the therapeutic dose by calculating absorbed doses in the tumor for efficacy and target organs for toxicity. It may be possible for PET imaging with specific tracers to evaluate therapeutic efficacy by measuring the amount of target molecules; however, the expression of the target molecules may change after the therapy—then, accurate response evaluation would be difficult with these target-specific PET studies.
Table 1. Representative Pair of Radiopharmaceuticals for Theranostics.
| Radiopharmaceutical for Therapy | Radiation | Half-Life | Radiopharmaceutical for Diagnosis |
|---|---|---|---|
| 177Lu-DOTA-TATE | Beta ray (β− particle) | 78 h | 68Ga-DOTA-TATE |
| 213Bi-DOTA-TOC | Alpha ray (He2+ particle) | 0.76 h | 68Ga-DOTA-TOC |
| 177Lu-PSMA | Beta ray (β− particle) | 78 h | 68Ga-PSMA |
| 225Ac-PSMA | Alpha ray (He2+ particle) | 10 d | 68Ga-PSMA |
DOTA-TATE: dodecane-tetraacetic acid-Tyr3-octreotate; DOTA-TOC: dodecane-tetraacetic acid-D-Phe1-Tyr3-octreotide; PSMA: prostate-specific membrane antigen.
Considering the present availability and required standardization, FDG-PET may be favorable for response evaluation in solid tumors. Since there is a variety of therapeutics that have effects on both cancer cells and the immune system, individualized evaluation criteria based on therapeutic agents and clinicopathologic information may be appropriate. Clinicopathologic data include therapeutic regimen and time from administration, immune function status, temporal changes in size and attenuation of tumor on CT, and pathological parameters, such as proliferation, invasion, differentiation, vascularity, and interstitial findings. These data as well as image features and quantitative indices like SUV and MTV of PET are subjected to artificial intelligence (AI) for radiomics analysis. Other available data such as MRI and contrast enhancement are welcome by AI for more detailed analyses.
Modalities used in the clinical setting include PET and single photon emission computed tomography, as well as MRI and ultrasonography. Optical imaging, such as fluorescence and bioluminescence imaging, plays an important role in preclinical settings; however, penetration of these signals is too shallow to detect labeled immune cells in clinical situations, and currently used contrast materials, such as gadolinium based agents, super paramagnetic iron oxide, and perfluorocarbon labeled with fluorine-19, for MRI are non-specific for immune cells. Therefore, nuclear medicine imaging is a possible procedure to elucidate anti-cancer immune responses [45][46]. Cell tracking of particular cell subsets would be done by radiolabeling in vitro prior to re-administration or by injecting a radiopharmaceutical that binds to a specific membrane antigen in vivo [47][48]. There have been many radiopharmaceuticals for cell tracking; however, none of these have been successfully used in clinical practice so far (Table 2).
Table 2. Potential Radiopharmaceuticals to Image Immune Cells and Cell Tracking.
| Target | Radiolabeling Agent | Application/Mechanism | References |
|---|---|---|---|
| T lymphocytes | 111In-oxine, 89Zr-oxine | Tumor infiltration | [46][49][50] |
| 18F-FDG | Cytokine production | ||
| SPIO | |||
| NK cells | 111In-oxine, 89Zr-oxine | Tumor infiltration | [46][51][52] |
| 18F-FDG, 11C-methyl iodide | NK cell homing | ||
| SPIO | |||
| Macrophages | 111In-oxine, 89Zr-nanoparticles | Tumor infiltration | [46][53][54][55][56][57] |
| 18F-FDG | Tumor-associated macrophages | ||
| SPIO, 19F-perfluorocarbon | |||
| Interleukin-2 | Iodine-123, Technetium-99 m, Fluorine-18 | Interleukin-2 receptors on T cells | [58] |
| Anti-CD8 cys-diabody | Zirconium-89, Copper-64 | CD8+ T cells | [59][60][61] |
| Anti-CD8 mAb | |||
| PK11195 | Carbon-11 | Tumor-associated macrophages, Translocator protein | [62] |
| Anti-TCR mAb | Copper-64 | Tumor infiltration of T cells | [63] |
| Anti-CD56 mAb | Technetium-99 m | NK cells | [64] |
18F-FDG: 2-[18F] fluoro-2-deoxy-d-glucose; SPIO: super paramagnetic iron oxide; TCR: T cell receptor; mAb: monoclonal antibody.
4. Conclusions
Quantitative parameters of FDG-PET, such as SUV and TLG, have been used to evaluate therapeutic responses. Metabolic changes and temporal enlargement due to immune cell infiltration seen after immune checkpoint inhibitors, anti-PD-1, and anti-PD-L1 antibodies facilitate the modification of FDG-PET response evaluation criteria as well as conventional RECIST. Dynamic interaction between cancer and immune cells, CSCs, and metabolism of cancer cells in the tumor microenvironment are promising targets to eradicate cancer. Accumulation of FDG reflects glucose metabolism of both cancer cells and immunologically competent cells in the tumor. Considering inter- and intra-patient tumor heterogeneity, immunological reaction to the therapy differs among patients according to the individual immune function and tumor heterogeneity. This limits the use of current response evaluation criteria and the revised ones may not be relevant enough for use in the clinical setting. Then, imaging of immune cells tracking may be crucial but is still a challenge, due to the fact that radiopharmaceuticals or MRI probes which are highly specific for biomarkers expressed in different immune cells are not likely to be determined. A radiomics approach which combines quantitative image features and deep learning algorithms has the potential to improve response assessment on the basis of elucidating pathologic mechanisms in more personalized treatments in the era of precision nuclear medicine. Multimodal imaging to highlight new therapeutic biomarkers in the complexed tumor response may be required to improve the management of cancer patients.
References
- Oriuchi, N.; Higuchi, T.; Ishikita, T.; Miyakubo, M.; Hanaoka, H.; Iida, Y.; Endo, K. Present role and future prospect of positron emission tomography in clinical oncology. Cancer Sci. 2006, 97, 1291–1297.
- Kitagawa, Y.; Sano, K.; Nishizawa, S.; Nakamura, M.; Ogasawara, T.; Sadato, N.; Yonekura, Y. FDG-PET for prediction of tumour aggressiveness and response to intra-arterial chemotherapy and radiotherapy in head and neck cancer. Eur. J. Nucl. Med. Mol. Imaging 2003, 30, 63–71.
- Warburg, O. The metabolism of carcinoma cells. J. Cancer Res. 1925, 9, 148–163. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218.
- Kernstine, K.H.; Faubert, B.; Do, Q.N.; Rogers, T.D.; Hensley, C.T.; Cai, J.; Torrealba, J.; Oliver, D.; Wachsmann, J.W.; Lenkinski, R.E.; et al. Does tumor FDG-PET avidity represent enhanced glycolytic metabolism in non-small cell lung cancer? Ann. Thorac. Surg. 2020, 109, 1019–1025.
- Momcilovic, M.; Jones, A.; Bailey, S.T.; Waldmann, C.M.; Li, R.; Lee, J.T.; Abdelhady, G.; Gomez, A.; Holloway, T.; Schmid, E.; et al. In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature 2019, 575, 380–384.
- DeBerardinis, R.J.; Navdeep, S.; Chandel, N.S. We need to talk about the Warburg effect. Nat. Metab. 2020, 2, 127–129.
- Hundshammer, C.; Braeuer, M.; Müller, C.A.; Hansen, A.E.; Schillmaier, M.; Düwel, S.; Feuerecker, B.; Glaser, S.J.; Haase, A.; Weichert, W.; et al. Simultaneous characterization of tumor cellularity and the Warburg effect with PET, MRI and hyperpolarized 13C-MRSI. Theranostics 2018, 8, 4765–4780.
- Ji, Z.; Long, H.; Hu, Y.; Qiu, X.; Chen, X.; Li, Z.; Fan, D.; Ma, B.; Fan, Q. Expression of MDR1, HIF-1α and MRP1 in sacral chordoma and chordoma cell line CM-319. J. Exp. Clin. Cancer Res. 2010, 29, 158.
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70.
- Vaupel, P.; Kallinowski, F.; Okunieff, P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: A review. Cancer Res. 1989, 49, 6449–6465.
- Brown, J.M.; Wilson, W.R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 2004, 4, 437–447.
- Cairns, R.A.; Mak, T.W. The current state of cancer metabolism. Nat. Rev. Cancer 2016, 16, 613–614.
- LaGory, E.L.; Giaccia, A.J. The ever-expanding role of HIF in tumour and stromal biology. Nat. Cell Biol. 2016, 18, 356–365.
- Chaffer, C.L.; San Juan, B.P.; Lim, E.; Weinberg, R.A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016, 35, 645–654.
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468.
- Liao, D.; Johnson, R.S. Hypoxia: A key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007, 26, 281–290.
- Kaira, K.; Endo, M.; Abe, M.; Nakagawa, K.; Ohde, Y.; Okumura, T.; Takahashi, T.; Murakami, H.; Tsuya, A.; Nakamura, Y.; et al. Biologic correlation of 2-[18F]-fluoro-2-deoxy-d-glucose uptake on positron emission tomography in thymic epithelial tumors. J. Clin. Oncol. 2010, 28, 3746–3753.
- Nagao, A.; Kobayashi, M.; Koyasu, S.; Chow, C.C.T.; Harada, H. HIF-1-Dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int. J. Mol. Sci. 2019, 20, 238.
- Tanaka, F.; Takenaka, K.; Oyanagi, H.; Fujinaga, T.; Otake, Y.; Yanagihara, K.; Ito, H.; Wada, H. Skip mediastinal nodal metastases in non-small cell lung cancer. Eur. J. Cardio-Thorac. Surg. 2004, 25, 1114–1120.
- Wang, H.; Zhou, Z.; Li, Y.; Chen, Z.; Lu, P.; Wang, W.; Liu, W.; Yu, L. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. 2017, 7, 11.
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine learning for medical imaging. Radiographics 2017, 37, 505–515.
- Endoh, H.; Yamamoto, R.; Ichikawa, A.; Shiozawa, S.; Nishizawa, N.; Satoh, Y.; Oriuchi, N. Clinicopathological significance of false-positive lymph node status on 18F-FDG PET in lung cancer. Clin. Lung Cancer 2020.
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Do, S.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current applications and future impact of machine learning in radiology. Radiology 2018, 288, 318–328.
- Betancur, J.; Rubeaux, M.; Fuchs, T.A.; Otaki, Y.; Arnson, Y.; Slipczuk, L.; Benz, D.C.; Germano, G.; Dey, D.; Lin, C.J.; et al. Automatic valve plane localization in myocardial perfusion SPECT/CT by machine learning: Anatomic and clinical validation. J. Nucl. Med. 2017, 58, 961–967.
- Kolossváry, M.; Park, J.; Bang, J.I.; Zhang, J.; Lee, J.M.; Paeng, J.C.; Merkely, B.; Narula, J.; Kubo, T.; Akasaka, T.; et al. Identification of invasive and radionuclide imaging markers of coronary plaque vulnerability using radiomic analysis of coronary computed tomography angiography. Eur. Heart J. Cardiovasc. Imaging 2019, 20, 1250–1258.
- Rao, V.M.; Levin, D.C.; Parker, L.; Cavanaugh, B.; Frangos, A.J.; Sunshine, J.H. How widely is computer-aided detection used in screening and diagnostic mammography? J. Am. Coll. Radiol. 2010, 7, 802–805.
- Fenton, J.J.; Abraham, L.; Taplin, S.H.; Geller, B.M.; Carney, P.A.; D’Orsi, C.; Elmore, J.G.; Barlow, W.E. Breast cancer surveillance consortium. effectiveness of computer-aided detection in community mammography practice. J. Natl. Cancer Inst. 2011, 103, 1152–1161.
- Mezrich, J.L.; Siegel, E.L. Legal ramifications of computer-aided detection in mammography. J. Am. Coll. Radiol. 2015, 12, 572–574.
- Halabi, S.S.; Prevedello, L.M.; Kalpathy-Cramer, J.; Mamonov, A.B.; Bilbily, A.; Cicero, M.; Pan, I.; Araújo Pereira, L.; Sousa, R.T.; Abdala, N.; et al. The RSNA pediatric bone age machine learning challenge. Radiology 2019, 290, 498–503.
- Chan, S.; Siegel, E.L. Will machine learning end the viability of radiology as a thriving medical specialty? Br. J. Radiol. 2019, 92, 20180416.
- Public Workshop—Evolving Role of Artificial Intelligence in Radiological Imaging; Comments of the American Colloge of Radiology. Available online: https://www.acr.org/-/media/ACR/NOINDEX/Advocacy/acr rsna comments fda-ai-evolvingrole-ws 6-30-2020.pdf (accessed on 20 August 2020).
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006.
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuzé, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018, 78, 4786–4789.
- Orlhac, F.; Nioche, C.; Soussan, M.; Buvat, I. Understanding changes in tumor texture indices in PET: A comparison between visual assessment and index values in simulated and patient data. J. Nucl. Med. 2017, 58, 387–392.
- Pyka, T.; Gempt, J.; Hiob, D.; Ringel, F.; Schlegel, J.; Bette, S.; Wester, H.J.; Meyer, B.; Förster, S. Textural analysis of pre-therapeutic [18F]-FET-PET and its correlation with tumor grade and patient survival in high-grade gliomas. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 133–141.
- Hatt, M.; Tixier, F.; Visvikis, D. Cheze le rest C. Radiomics in PET/CT: More than meets the eye? J. Nucl. Med. 2017, 58, 365–366.
- Fornacon-Wood, I.; Faivre-Finn, C.; O’Connor, J.P.B.; Price, G.J. Radiomics as a personalized medicine tool in lung cancer: Separating the hope from the hype. Lung Cancer 2020, 146, 197–208.
- Sunaga, N.; Oriuchi, N.; Kaira, K.; Yanagitani, N.; Tomizawa, Y.; Hisada, T.; Ishizuka, T.; Endo, K.; Mori, M. Usefulness of FDG-PET for early prediction of the response to gefitinib in non-small cell lung cancer. Lung Cancer 2008, 59, 203–210.
- Kaira, K.; Higuchi, T.; Naruse, I.; Arisaka, Y.; Tokue, A.; Altan, B.; Suda, S.; Mogi, A.; Shimizu, K.; Sunaga, N.; et al. Metabolic activity by 18F-FDG-PET/CT is predictive of early response after nivolumab in previously treated NSCLC. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 56–66.
- Shimizu, K.; Kaira, K.; Higuchi, T.; Hisada, T.; Yokobori, T.; Oyama, T.; Asao, T.; Tsushima, Y.; Shirabe, K. Relationship between tumor immune markers and Fluorine-18-α-Methyltyrosine ([18F]FAMT) uptake in PAtients with Lung Cancer. Mol. Imaging Biol. 2020, 22, 1078–1086.
- Nishino, M.; Jagannathan, J.P.; Krajewski, K.M.; O’Regan, K.; Hatabu, H.; Shapiro, G.; Ramaiya, N.H. Personalized tumor response assessment in the era of molecular medicine: Cancer-Specific and therapy-specific response criteria to complement pitfalls of RECIST. AJR Am. J. Roentgenol. 2012, 198, 737–745.
- Suzuki, S.; Kaira, K.; Ohshima, Y.; Ishioka, N.S.; Sohda, M.; Yokobori, T.; Miyazaki, T.; Oriuchi, N.; Tominaga, H.; Kanai, Y.; et al. Biological significance of fluorine-18-α-methyltyrosine (FAMT) uptake on PET in patients with oesophageal cancer. Br. J. Cancer 2014, 110, 1985–1991.
- Kaira, K.; Higuchi, T.; Sunaga, N.; Arisaka, Y.; Hisada, T.; Tominaga, H.; Oriuchi, N.; Asao, T.; Tsushima, Y.; Yamada, M. Usefulness of 18F-α-Methyltyrosine PET for therapeutic monitoring of patients with advanced Lung Cancer. Anti-Cancer Res. 2016, 36, 6481–6490.
- Ehlerding, E.B.; England, C.G.; McNeel, D.G.; Cai, W. Molecular imaging of immunotherapy targets in cancer. J. Nucl. Med. 2016, 57, 1487–1492.
- Ahrens, E.T.; Bulte, J.W. Tracking immune cells in vivo using magnetic resonance imaging. Nat. Rev. Immunol. 2013, 13, 755–763.
- Yongtao, Z.; Jiongwei, H.; Tongming, Z.; Ronggang, L.; Zhifu, W.; Fukai, M.; Jianhong, Z. Stem Cell Tracking Technologies for Neurological Regenerative Medicine Purposes. Stem Cells Int. 2017, 2017, 2934149.
- Zeelen, C.; Paus, C.; Draper, D.; Heskamp, S.; Signore, A.; Galli, F.; Griessinger, C.M.; Aarntzen, E.H. In vivo imaging of tumor-infiltrating immune cells: Implications for cancer immunotherapy. Q. J. Nucl. Med. Mol. Imaging 2018, 62, 56–77.
- Ritchie, D.; Mileshkin, L.; Wall, D.; Bartholeyns, J.; Thompson, M.; Coverdale, J.; Lau, E.; Wong, J.; Eu, P.; Hicks, R.J.; et al. In vivo tracking of macrophage activated killer cells to sites of metastatic ovarian carcinoma. Cancer Immunol. Immunother. 2007, 56, 155–163.
- Bansal, A.; Pandey, M.K.; Demirhan, Y.E.; Nesbitt, J.J.; Crespo-Diaz, R.J.; Terzic, A.; Behfar, A.; DeGrado, T.R. Novel 89Zr cell labeling approach for PET-based cell trafficking studies. EJNMMI Res. 2015, 5, 19.
- Meller, B.; Frohn, C.; Brand, J.M.; Lauer, I.; Schelper, L.F.; von Hof, K.; Kirchner, H.; Richter, E.; Baehre, M. Monitoring of a new approach of immunotherapy with allogenic 111In-labelled NK cells in patients with renal cell carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2004, 31, 403–407.
- Melder, R.J.; Brownell, A.L.; Shoup, T.M.; Brownell, G.L.; Jain, R.K. Imaging of activated natural killer cells in mice by positron emission tomography: Preferential uptake in tumors. Cancer Res. 1993, 53, 5867–5871.
- Quillien, V.; Moisan, A.; Lesimple, T.; Leberre, C.; Toujas, L. Biodistribution of 111indium-labeled macrophages infused intravenously in patients with renal carcinoma. Cancer Immunol. Immunother. 2001, 50, 477–482.
- Weissleder, R.; Nahrendorf, M.; Pittet, M.J. Imaging macrophages with nanoparticles. Nat. Mater. 2014, 13, 125–138.
- Srinivas, M.; Boehm-Sturm, P.; Figdor, C.G.; de Vries, I.J.; Hoehn, M. Labeling cells for in vivo tracking using 19F MRI. Biomaterials 2012, 33, 8830–8840.
- Pérez-Medina, C.; Tang, J.; Abdel-Atti, D.; Hogstad, B.; Merad, M.; Fisher, E.A.; Fayad, Z.A.; Lewis, J.S.; Mulder, W.J.; Reiner, T. PET Imaging of Tumor-Associated Macrophages with 89Zr-Labeled High-Density Lipoprotein Nanoparticles. J. Nucl. Med. 2015, 56, 1272–1277.
- Schniering, J.; Benešová, M.; Brunner, M.; Haller, S.; Cohrs, S.; Frauenfelder, T.; Vrugt, B.; Feghali-Bostwick, C.; Schibli, R.; Distler, O.; et al. 18F-AzaFol for Detection of Folate Receptor-β Positive Macrophages in Experimental Interstitial Lung Disease-A Proof-of-Concept Study. Front. Immunol. 2019, 10, 2724.
- Markovic, S.N.; Galli, F.; Suman, V.J.; Nevala, W.K.; Paulsen, A.M.; Hung, J.C.; Gansen, D.N.; Erickson, L.A.; Marchetti, P.; Wiseman, G.A.; et al. Non-invasive visualization of tumor infiltrating lymphocytes in patients with metastatic melanoma undergoing immune checkpoint inhibitor therapy: A pilot study. Oncotarget 2018, 9, 30268–30278.
- Tavaré, R.; McCracken, M.N.; Zettlitz, K.A.; Salazar, F.B.; Olafsen, T.; Witte, O.N.; Wu, A.M. Immuno-PET of Murine T Cell Reconstitution Postadoptive Stem Cell Transplantation Using Anti-CD4 and Anti-CD8 Cys-Diabodies. J. Nucl. Med. 2015, 56, 1258–1264.
- Tavaré, R.; Escuin-Ordinas, H.; Mok, S.; McCracken, M.N.; Zettlitz, K.A.; Salazar, F.B.; Witte, O.N.; Ribas, A.; Wu, A.M. An Effective Immuno-PET imaging method to monitor CD8-Dependent responses to immunotherapy. Cancer Res. 2016, 76, 73–82.
- Seo, J.W.; Tavaré, R.; Mahakian, L.M.; Silvestrini, M.T.; Tam, S.; Ingham, E.S.; Salazar, F.B.; Borowsky, A.D.; Wu, A.M.; Ferrara, K.W. CD8+ T-Cell Density Imaging with 64Cu-Labeled Cys-Diabody Informs Immunotherapy Protocols. Clin. Cancer Res. 2018, 24, 4976–4987.
- Zheng, J.; Boisgard, R.; Siquier-Pernet, K.; Decaudin, D.; Dollé, F.; Tavitian, B. Differential expression of the 18 kDa translocator protein (TSPO) by neoplastic and inflammatory cells in mouse tumors of breast cancer. Mol. Pharm 2011, 8, 823–832.
- Griessinger, C.; Maurer, A.; Kesenheimer, C.; Kehlbach, R.; Reischl, G.; Ehrlichmann, W.; Bukala, D.; Harant, M.; Cay, F.; Brück, J.; et al. 64Cu antibody-targeting of the T cell receptor and subsequent internalization enables in vivo tracking of lymphocytes by PET. Proc. Natl. Acad. Sci. USA 2015, 112, 1161–1166.
- Galli, F.; Brück, J.; Rapisarda, A.S.; Stabile, H.; Malviya, G.; Manni, I.; Bonanno, E.; Piaggio, G.; Gismondi, A.; Santoni, A.; et al. In vivo imaging of natural killer cell trafficking in tumors. J. Nucl. Med. 2015, 56, 1575–1580.




