
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Chaiyavat Chaiyasut | -- | 3343 | 2022-08-05 05:57:31 | | | |
| 2 | Sirius Huang | -1 word(s) | 3342 | 2022-08-05 07:47:31 | | |
Video Upload Options
Parkinson’s disease (PD) is the second-most prevalent neurodegenerative or neuropsychiatric disease, affecting 1% of seniors worldwide. The gut microbiota (GM) is one of the key access controls for most diseases and disorders. Disturbance in the GM creates an imbalance in the function and circulation of metabolites, resulting in unhealthy conditions. Any dysbiosis could affect the function of the gut, consequently disturbing the equilibrium in the intestine, and provoking pro-inflammatory conditions in the gut lumen, which send signals to the central nervous system (CNS) through the vagus enteric nervous system, possibly disturbing the blood–brain barrier. The neuroinflammatory conditions in the brain cause accumulation of α-syn, and progressively develop PD. An important aspect of understanding and treating the disease is access to broad knowledge about the influence of dietary supplements on GM.
1. Introduction
1.1. Gut Microbiota
1.2. Gut Microbiota, Neuronal System, and Neurodegenerative Diseases (NDs)
1.3. Parkinson’s Disease (PD)
2. Pathophysiology of Parkinson’s Disease
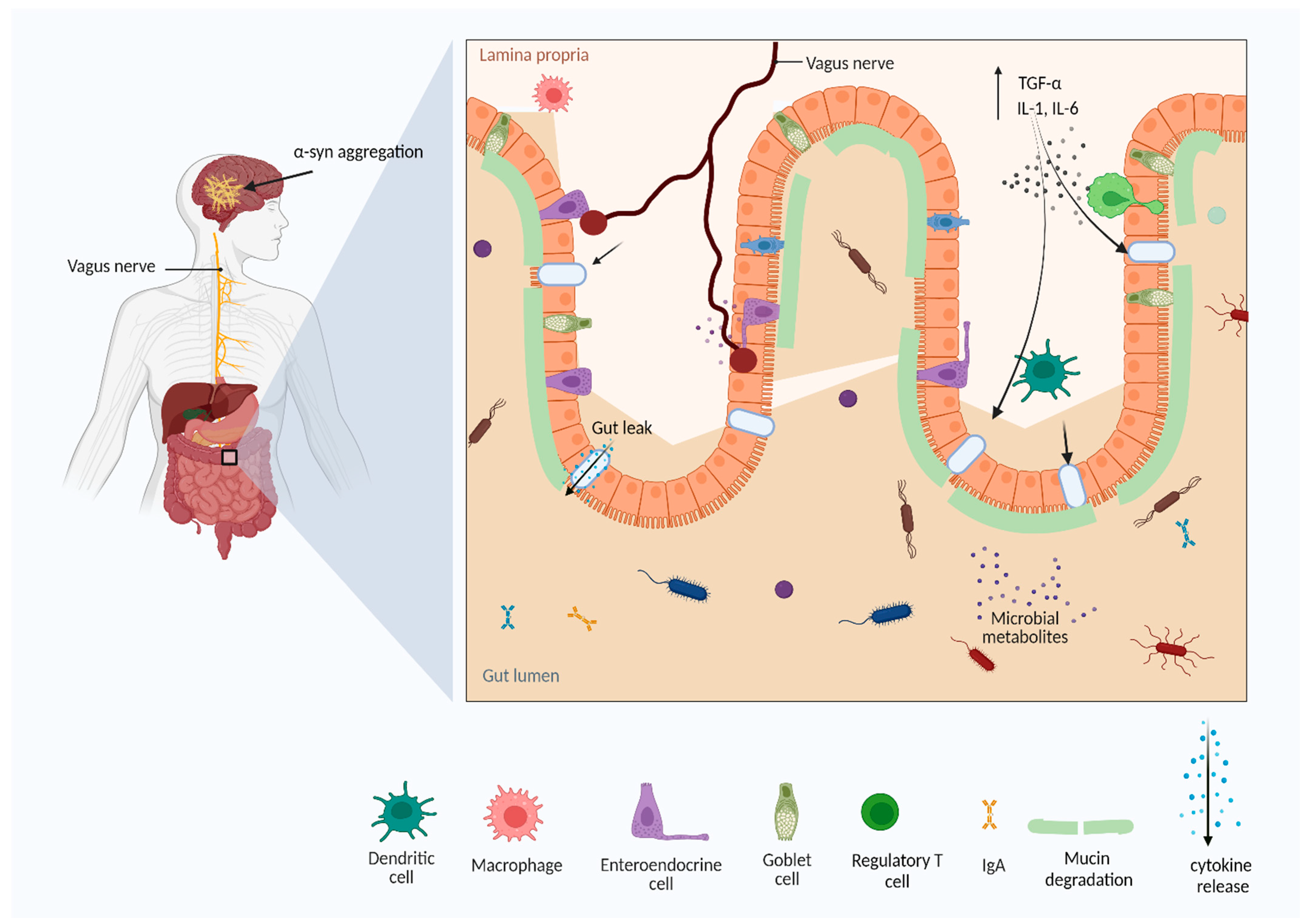
2.1. Toll-Like Receptors Expression in PD
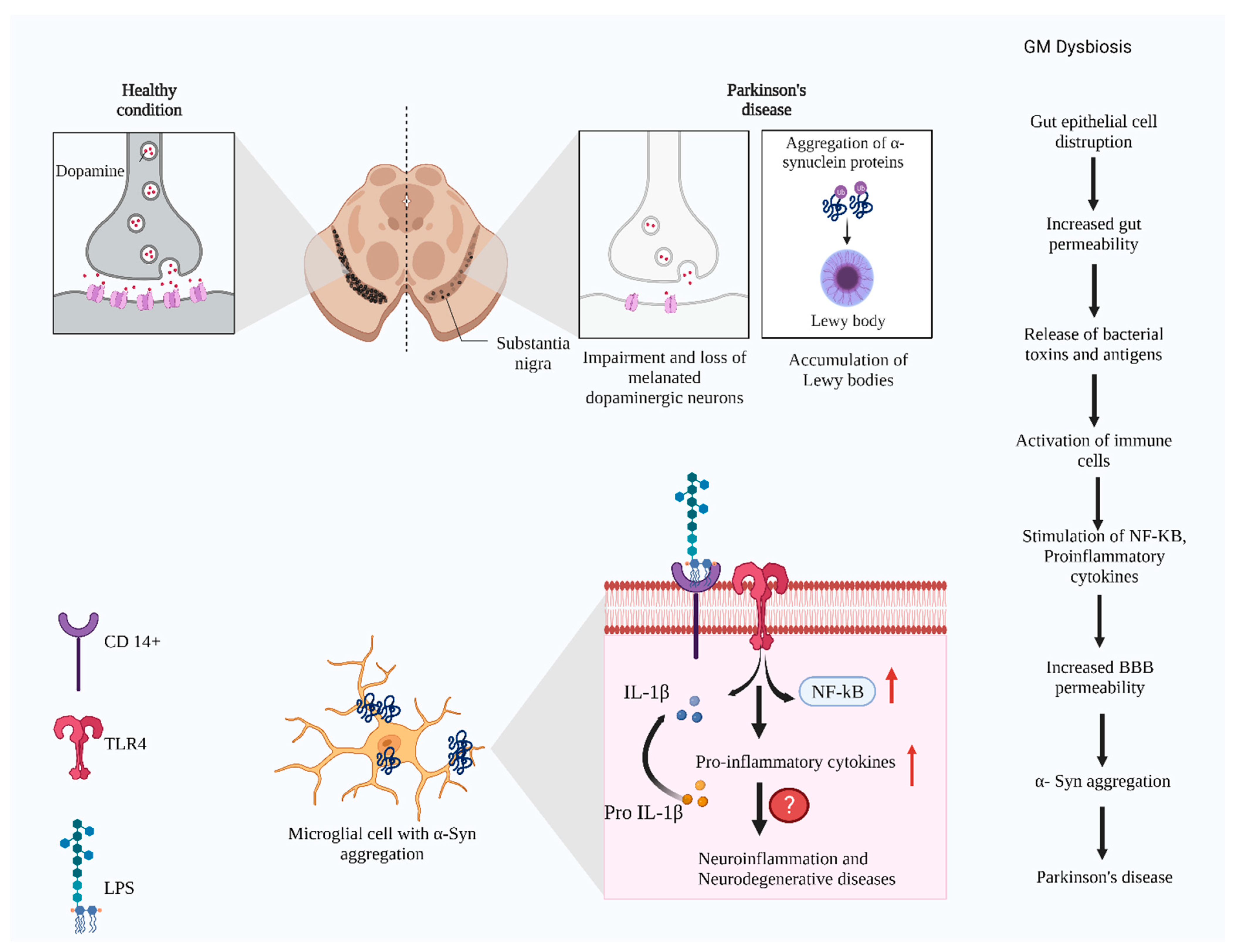
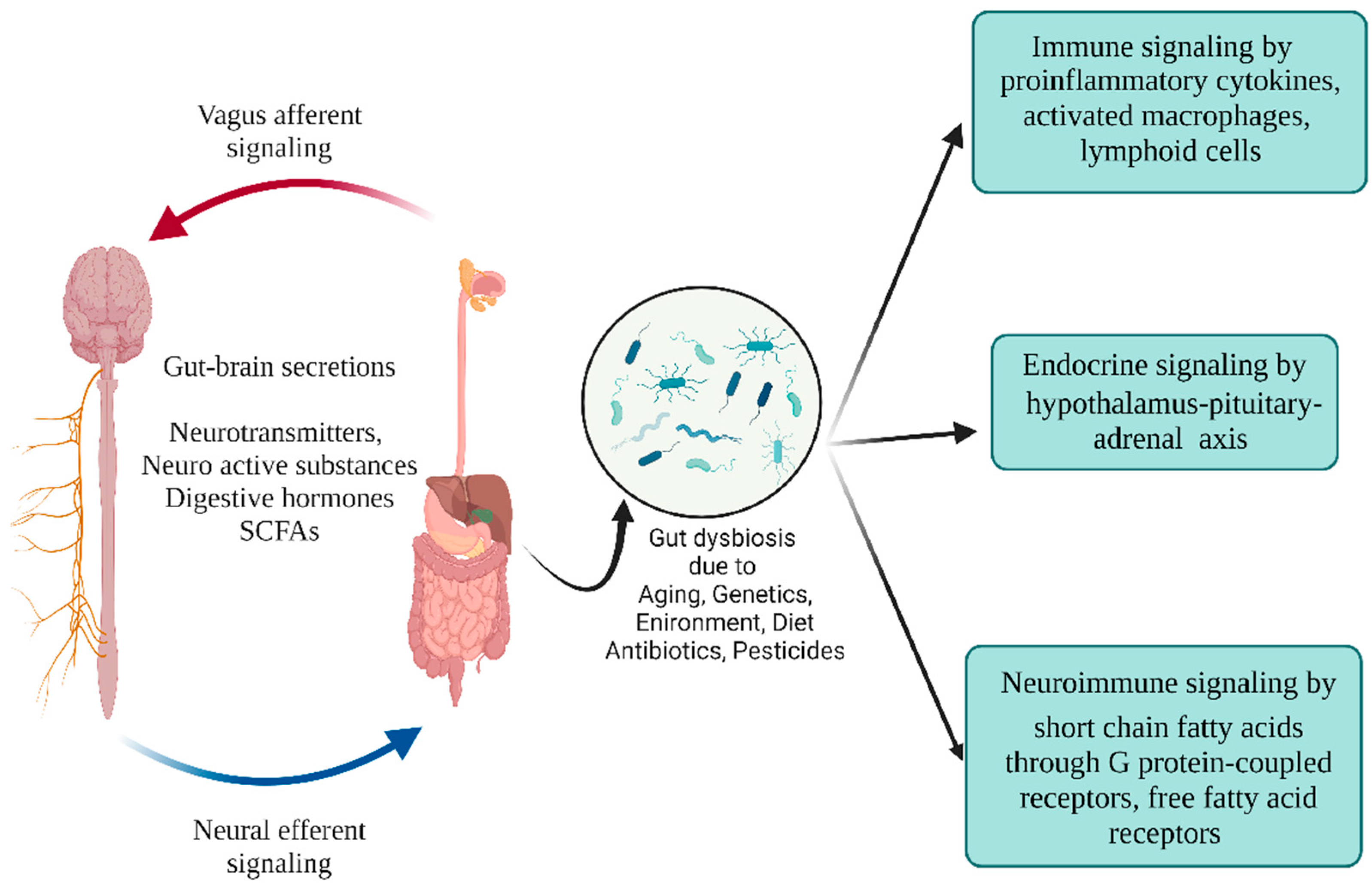
3. Signaling Pathways Associated with Gut Microbial Changes and PD
4. Gut Microbiota and Parkinson’s Disease
References
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.; Sandhu, K.V.; Bastiaanssen, T.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013.
- Ding, R.X.; Goh, W.R.; Wu, R.N.; Yue, X.Q.; Luo, X.; Khine, W.; Wu, J.R.; Lee, Y.K. Revisit gut microbiota and its impact on human health and disease. J. Food Drug Anal. 2019, 27, 623–631.
- Kilia, M.; Chapple, I.L.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome-an update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666.
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379.
- Zhu, X.; Han, Y.; Du, J.; Liu, R.; Jin, K.; Yi, W. Microbiota-gut-brain axis and the central nervous system. Oncotarget 2017, 8, 53829–53838.
- Schnabl, B.; Brenner, D.A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014, 146, 1513–1524.
- Lazar, V.; Ditu, L.M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front. Immunol. 2018, 2018 9, 1830.
- Sartor, R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008, 134, 577–594.
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65.
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638.
- Rajilic-Stojanovic, M.; Smidt, H.; de Vos, W.M. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 2007, 9, 2125–2136.
- Lin, C.S.; Chang, C.J.; Lu, C.C.; Martel, J.; Ojcius, D.M.; Ko, Y.F.; Young, J.D.; Lai, H.C. Impact of the gut microbiota, prebiotics, and probiotics on human health and disease. Biomed. J. 2014, 37, 259–268.
- Matricon, J.; Meleine, M.; Gelot, A.; Piche, T.; Dapoigny, M.; Muller, E.; Ardid, D. Associations between immune activation, intestinal permeability and the irritable bowel syndrome. Aliment. Pharmacol. Ther. 2012, 36, 1009–1031.
- Hughes, P.A.; Zola, H.; Penttila, I.A.; Blackshaw, L.A.; Andrews, J.M.; Krumbiegel, D. Immune activation in irritable bowel syndrome: Can neuroimmune interactions explain symptoms? Am. J. Gastroenterol. 2013, 108, 1066–1074.
- Forsythe, P.; Kunze, W.A. Voices from within gut microbes and the CNS. Cell Mol. Life Sci. 2013, 70, 55–69.
- Luczynski, P.; Tramullas, M.; Viola, M.; Shanahan, F.; Clarke, G.; O’Mahony, S.; Dinan, T.G.; Cryan, J.F. Microbiota regulates visceral pain in the mouse. eLife 2017, 6, e25887.
- Foster, J.A.; Lyte, M.; Meyer, E.; Cryan, J.F. Gut microbiota and brain function: An evolving field in neuroscience. Int. J. Neuropsychopharmacol. 2016, 19, pyv114.
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712.
- Dinan, T.G.; Cryan, J.F. Regulation of the stress response by the gut microbiota: Implications for psychoneuroendocrinology. Psychoneuroendocrinology 2012, 37, 1369–1378.
- Stilling, R.M.; Dinan, T.G.; Cryan, J.F. Microbial genes, brain & behavior—Epigenetic regulation of the gut-brain axis. Genes Brain Behav. 2014, 13, 69–86.
- Obata, Y.; Pachnis, V. The Effect of Microbiota and the Immune System on the Development and Organization of the Enteric Nervous System. Gastroenterology 2016, 151, 836–844.
- Rao, M.; Gershon, M.D. The bowel and beyond: The enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 517.
- Westfall, S.; Lomis, N.; Kahouli, I.; Dia, S.Y.; Singh, S.P.; Prakash, S. Microbiome, probiotics and neurodegenerative diseases: Deciphering the gut brain axis. Cell. Mol. Life Sci. 2017, 74, 3769–3787.
- Rhee, S.H.; Pothoulakis, C.; Mayer, E.A. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat. Rev. Gastroenterol Hepatol. 2009, 6, 306–314.
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 2018, 76, 473–493.
- Burger-van Paassen, N.; Vincent, A.; Puiman, P.J.; van der Sluis, M.; Bouma, J.; Boehm, G.; van Goudoever, J.B.; van Seuningen, I.; Renes, I.B. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: Implications for epithelial protection. Biochem. J. 2009, 420, 211–219.
- Nell, S.; Suerbaum, S.; Josenhans, C. The impact of the microbiota on the pathogenesis of IBD: Lessons from mouse infection models. Nat. Rev. Microbiol. 2010, 8, 564–577.
- Doroszkiewicz, J.; Groblewska, M.; Mroczko, B. The Role of Gut Microbiota and Gut-Brain Interplay in Selected Diseases of the Central Nervous System. Int. J. Mol. Sci. 2021, 22, 10028.
- Kelly, J.R.; Borre, Y.; O’Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118.
- De Palma, G.; Lynch, M.D.; Lu, J.; Dang, V.T.; Deng, Y.; Jury, J.; Umeh, G.; Miranda, P.M.; Pigrau Pastor, M.; Sidani, S.; et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 2017, 9, eaaf6397.
- Gentile, F.; Doneddu, P.E.; Riva, N.; Nobile-Orazio, E.; Quattrini, A. Diet, Microbiota and Brain Health: Unravelling the Network Intersecting Metabolism and Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 7471.
- Diaz Heijtz, R.; Wangm, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052.
- Ogbonnaya, E.S.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; O’Leary, O.F. Adult Hippocampal Neurogenesis Is Regulated by the Microbiome. Biol. Psychiatry 2015, 78, e7–e9.
- Rutsch, A.; Kantsjö, J.B.; Ronchi, F. The Gut-Brain Axis: How Microbiota and Host Inflammasome Influence Brain Physiology and Pathology. Front. Immunol. 2020, 11, 604179.
- Shen, T.; Yue, Y.; He, T.; Huang, C.; Qu, B.; Lv, W.; Lai, H.Y. The Association Between the Gut Microbiota and Parkinson’s Disease, a Meta-Analysis. Front. Aging Neuros. 2021, 13, 636545.
- Martinez-Martin, P. The importance of non-motor disturbances to quality of life in Parkinson’s disease. J. Neurol. Sci. 2011, 310, 12–16.
- Su, A.; Gandhy, R.; Barlow, C.; Triadafilopoulos, G. A practical review of gastrointestinal manifestations in Parkinson’s disease. Parkinsonism Relat. Disord. 2017, 39, 17–26.
- Lang, A.E. A critical appraisal of the premotor symptoms of Parkinson’s disease: Potential usefulness in early diagnosis and design of neuroprotective trials. Mov. Disord. 2011, 26, 775–783.
- Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis in health and disease. Gastroenterol. Clin. N. Am. 2017, 46, 77–89.
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184.
- Myers, S.P.; Hawrelak, J. The causes of intestinal dysbiosis: A review. Altern. Med. Rev. 2004, 9, 180–197.
- Rietdijk, C.D.; Perez-Pardo, P.; Garssen, J.; Van Wezel, R.J.; Kraneveld, A.D. Exploring braak’s hypothesis of Parkinson’s disease. Front. Neurol. 2017, 8, 37.
- Parkinson, J. An essay on the shaking palsy. 1817. J. Neuropsychiatry Clin. Neurosci. 2002, 14, 222–223.
- Bjorklund, A.; Dunnett, S.B. Dopamine neuron systems in the brain: An update. Trends Neurosci. 2007, 30, 194–202.
- Hornykiewicz, O. The discovery of dopamine deficiency in the parkinsonian brain. J. Neural Transm. 2006, 70, 9–15.
- McNaught, K.S.P.; Jenner, P.; Olanow, C.W. Protein mishandling: Role of the ubiquitin-proteasome system in the pathogenesis of Parkinson’s disease. In Parkinson’s Disease and Movement Disorders, 6th ed.; Jankovic, J., Tolosa, E., Eds.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2007; pp. 33–49.
- Pan, T.; Kondo, S.; Le, W.; Jankovic, J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson’s disease. Brain 2008, 131, 1969–1978.
- Braak, H.; Müller, C.M.; Rüb, U.; Ackermann, H.; Bratzke, H.; de Vos, R.A.; Del Tredici, K. Pathology associated with sporadic Parkinson’s disease—Where does it end? J. Neural Transm. Suppl. 2006, 70, 89–97.
- Galvan, A.; Wichmann, T. Pathophysiology of parkinsonism. Clin. Neurophysiol. 2008, 119, 1459–1474.
- Brook, D.J. Imaging of genetic and degenerative disorders primarily causing Parkinsonism. Handb. Clin. Neurol. 2016, 135, 493–505.
- Bentea, E.; Verbruggen, L.; Massie, A. The proteosome inhibition model of Parkinson’s disease. J. Parkinson Dis. 2017, 7, 31–63.
- Bishop, P.; Rocca, D.; Henley, J.M. Ubiquitin C-terminal hydrolase L1 (UCH-L1): Structure, distribution and roles in brain function and dysfunction. Biochem. J. 2016, 473, 2453–2462.
- Htike, T.T.; Mishra, S.; Kumar, S.; Padmanabhan, P.; Gulyas, B. Peripheral biomarkers for early detection of Alzheimer’s disease and Parkinson’s diseases. Mol. Neurobiol. 2019, 56, 2256–2277.
- Burke, W.J.; Li, S.W.; Williams, E.A.; Nonneman, R.; Zam, D.S. 3,4-Dihydroxyphenylacetaldehyde is the toxic dopamine metabolite in vivo: Implications for Parkinson’s disease pathogenesis. Brain Res. 2003, 989, 205–213.
- Braak, H.; Rub, U.; Gai, W.; Del Tredici, K. Idiopathic Parkinson’s disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J. Neural. Transm. 2003, 110, 517–536.
- Beach, T.G.; Adler, C.H.; Sue, L.I.; Vedders, L.; Lue, L.; White Iii, C.L.; Akiyama, H.; Caviness, J.N.; Shill, H.A.; Sabbagh, M.N.; et al. Arizona Parkinson’s Disease Consortium. Multi-organ distribution of phosphorylated alpha-synuclein histopathology in subjects with Lewy body disorders. Acta Neuropathol. 2010, 119, 689–702.
- Ugrumov, M. Development of early diagnosis of Parkinson’s disease: Illusion or reality? CNS Neurosci. Ther. 2020, 26, 997–1009.
- Sun, Q.; Wang, S.; Chen, J.; Cai, H.; Huang, W.; Zhang, Y.; Wang, L.; Xing, Y. MicroRNA-190 alleviates neuronal damage and inhibits neuroinflammation via Nlrp3 in MPTP-induced Parkinson’s disease mouse model. J. Cell. Physiol. 2019, 234, 23379–23387.
- Delenclos, M.; Jones, D.R.; McLean, P.J.; Uitti, R.J. Biomarkers in Parkinson’s disease: Advances and strategies. Parkinsonism Relat. Disord. 2016, 22, S106–S110.
- Cowan, K.; Anichtchik, O.; Luo, S. Mitochondrial integrity in neurodegeneration. CNS Neurosci. Ther. 2019, 25, 825–836.
- Emamzadeh, F.N.; Surguchov, A. Parkinson’s disease: Biomarkers, treatment, and risk factors. Front. Neurosci. 2018, 12, 612.
- Cova, I.; Priori, A. Diagnostic biomarkers for Parkinson’s disease at a glance: Where are we? J. Neural Transm. 2018, 125, 1417–1432.
- Deng, H.; Wang, P.; Jankovic, J. The genetics of Parkinson disease. Ageing Res. Rev. 2018, 42, 72–85.
- He, R.; Yan, X.; Guo, J.; Xu, Q.; Tang, B.; Sun, Q. Recent advances in biomarkers for Parkinson’s disease. Front. Aging Neurosci. 2018, 10, 305.
- Blauwendraat, C.; Mike, A.; Nalls, M.A.; Andrew, B.; Singleton, A.B. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020, 19, 170–178.
- Licker, V.; Kövari, E.; Hochstrasser, D.F.; Burkhard, P.R. Proteomics in human Parkinson’s disease research. J. Proteomics 1997, 73, 10–29.
- Overk, C.R.; Masliah, E. Pathogenesis of synaptic degeneration in Alzheimer’s disease and Lewy body disease. Biochem. Pharmacol. 2014, 88, 508–516.
- Hirsch, E.C.; Vyas, S.; Hunot, S. Neuroinflammation in Parkinson’s disease. Parkinsonism Relat. Disord. 2012, 8, S210–S212.
- Mosley, R.L.; Hutter-Saunders, J.A.; Stone, D.K.; Gendelman, H.E. Inflammation and adaptive immunity in Parkinson’s disease. Cold. Spring Harb. Perspect. Med. 2012, 2, a009381.
- Jankovic, J. Pathophysiology and assessment of parkinsonian symptoms and signs. In Handbook of Parkinson’s Disease; Pahwa, R., Lyons, K., Koller, W.C., Eds.; Taylor and Francis Group: New York, NY, USA, 2007; pp. 79–104.
- Berardelli, A.; Rothwell, J.C.; Thompson, P.D.; Hallett, M. Pathophysiology of bradykinesia in Parkinson’s disease. Brain 2001, 124, 2131–2146.
- Ross, G.W.; Petrovitch, H.; Abbott, R.D.; Nelson, J.; Markesbery, W.; Davis, D.; Hardman, J.; Launer, L.; Masaki, K.; Tanner, C.M.; et al. Parkinsonian signs and substantia nigra neuron density in decendents elders without PD. Ann. Neurol. 2004, 56, 532–539.
- Shulman, L.M.; Singer, C.; Bean, J.A.; Weiner, W.J. Internal tremor in patients with Parkinson’s disease. Mov. Disord. 1996, 11, 3–7.
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376.
- Riley, D.; Lang, A.E.; Blair, R.D.; Birnbaum, A.; Reid, B. Frozen shoulder and other shoulder disturbances in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 1989, 52, 63–66.
- Stamey, W.P.; Jankovic, J. Shoulder pain in Parkinson’s disease. Mov. Disord. 2007, 22, S247–S248.
- Ashour, R.; Tintner, R.; Jankovic, J. Striatal deformities of the hand and foot in Parkinson’s disease. Lancet Neurol. 2005, 4, 423–431.
- Ashour, R.; Jankovic, J. Joint and skeletal deformities in Parkinson’s disease, multiple system atrophy, and progressive supranuclear palsy. Mov. Disord. 2006, 21, 1856–1863.
- Askmark, H.; Eeg-Olofsson, K.E.; Johansson, A.; Nilsson, P.; Olsson, Y.; Aquilonius, S. Parkinsonism and neck extensor myopathy: A new syndrome or coincidental findings? Arch. Neurol. 2001, 58, 232–237.
- Djaldetti, R.; Melamed, E. Camptocormia in Parkinson’s disease: New insights. J. Neurol. Neurosurg. Psychiatry 2006, 77, 1205.
- Azher, S.N.; Jankovic, J. Camptocormia: Pathogenesis, classification, and response to therapy. Neurology 2005, 65, 355–359.
- Williams, D.R.; Watt, H.C.; Lees, A.J. Predictors of falls and fractures in bradykinetic rigid syndromes: A retrospective study. J. Neurol. Neurosurg. Psychiatry 2006, 77, 468–473.
- Giladi, N.; McDermott, M.P.; Fahn, S.; Przedborski, S.; Jankovic, J.; Stern, M.; Tanner, C.; Parkinson Study Group. Freezing of gait in PD: Prospective assessment in the DATATOP cohort. Neurology 2001, 56, 1712–1721.
- Boghen, D. Apraxia of lid opening: A review. Neurology 1997, 48, 1491–1494.
- Macht, M.; Kaussner, Y.; Moller, J.C.; Stiasny-Kolster, K.; Eggert, K.M.; Krüger, H.P.; Ellgring, H. Predictors of freezing in Parkinson’s disease: A survey of 6620 patients. Mov. Disord. 2007, 22, 953–956.
- Aziz, Q.; Thompson, D.G. Brain-gut axis in health and disease. Gastroenterology 1998, 114, 559–578.
- Micieli, G.; Tosi, P.; Marcheselli, S.; Cavallini, A. Autonomic dysfunction in Parkinson’s disease. Neurol. Sci. 2003, 24 (Suppl. S1), S32–S34.
- Mulak, A.; Bonaz, B. Brain-gut-microbiota axis in Parkinson’s disease. World J. Gastroenterol. 2015, 21, 10609–10620.
- Braak, H.; Del Tredici, K.; Rub, U.; De Vos, R.A.I.; Steur, E.N.H.J.; Braak, E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 2003, 24, 197–211.
- Aderem, A.; Ulevitch, R.J. Toll-like receptors in the induction of the innate immune response. Nature 2000, 406, 782–787.
- Flaherty, S.; Reynolds, J.M. TLR Function in Murine CD4+ T Lymphocytes and Their Role in Inflammation. Methods Mol. Biol. 2016, 1390, 215–227.
- Liu, G.; Zhao, Y. Toll-like receptors and immune regulation: Their direct and indirect modulation on regulatory CD4+ CD25+ T cells. Immunology 2007, 122, 149–156.
- Campos-Acuna, J.; Elgueta, D.; Pacheco, R. T-cell-driven inflammation as a mediator of the gut-brain axis involved in Parkinson’s disease. Front. Immunol. 2019, 10, 239.
- Fujiwara, H.; Hasegawa, M.; Dohmae, N.; Kawashima, A.; Masliah, E.; Goldberg, M.S.; Shen, J.; Takio, K.; Iwatsubo, T. Alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002, 4, 160–164.
- Alvarez-Erviti, L.; Couch, Y.; Richardson, J.; Cooper, J.M.; Wood, M.J. Alpha-synuclein release by neurons activates the inflammatory response in a microglial cell line. J. Neurosci. Res. 2011, 69, 337–342.
- Béraud, D.; Twomey, M.; Bloom, B.; Mittereder, A.; Ton, V.; Neitzke, K.; Chasovskikh, S.; Mhyre, T.R.; Maguire-Zeiss, K.A. α-Synuclein Alters Toll-Like Receptor Expression. Front. Neurosci. 2011, 5, 80.
- Doorn, K.J.; Moors, T.; Drukarch, B.; van de Berg, W.; Lucassen, P.J.; van Dam, A.M. Microglial phenotypes and toll-like receptor 2 in the substantia nigra and hippocampus of incidental Lewy body disease cases and Parkinson’s disease patients. Acta Neuropathol. Commun. 2014, 2, 90.
- Soares, J.B.; Pimentel-Nunes, P.; Roncon-Albuquerque, R.; Leite-Moreira, A. The role of lipopolysaccharide/toll-like receptor 4 signaling in chronic liver diseases. Hepatol. Int. 2010, 4, 659–672.
- Letiembre, M.; Liu, Y.; Walter, S.; Hao, W.; Pfander, T.; Wrede, A.; Schulz-Schaeffer, W.; Fassbender, K. Screening of innate immune receptors in neurodegenerative diseases: A similar pattern. Neurobiol. Aging 2009, 30, 759–768.
- Azam, S.; Jakaria, M.; Kim, I.S.; Kim, J.; Haque, M.E.; Choi, D.K. Regulation of Toll-Like Receptor (TLR) Signaling Pathway by Polyphenols in the Treatment of Age-Linked Neurodegenerative Diseases: Focus on TLR4 Signaling. Front. Immunol. 2019, 10, 1000.
- Rietdijk, C.D.; Van Wezel, R.J.A.; Garssen, J.; Kraneveld, A.D. Neuronal toll-like receptors and neuro-immunity in Parkinson’s disease, Alzheimer’s disease and stroke. Neuroimmunol. Neuroinflamm. 2016, 3, 27–37.
- Kim, C.; Ho, D.H.; Suk, J.E.; You, S.; Michael, S.; Kang, J.; Joong Lee, S.; Masliah, E.; Hwang, D.; Lee, H.J.; et al. Neuron-released oligomeric α-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat. Commun. 2013, 4, 1562.
- Fellner, L.; Irschick, R.; Schanda, K.; Reindl, M.; Klimaschewski, L.; Poewe, W.; Wenning, G.K.; Stefanova, N. Toll-like receptor 4 is required for alpha-synuclein dependent activation of microglia and astroglia. Glia 2013, 61, 349–360.
- Kim, C.; Spencer, B.; Rockenstein, E.; Yamakado, H.; Mante, M.; Adame, A.; Fields, J.A.; Masliah, D.; Iba, M.; Lee, H.J.; et al. Immunotherapy targeting toll-like receptor 2 alleviates neurodegeneration in models of synucleinopathy by modulating α-synuclein transmission and neuroinflammation. Mol. Neurodegener. 2018, 13, 43.
- Venezia, S.; Refolo, V.; Polissidis, A.; Stefanis, L.; Wenning, G.K.; Stefanova, N. Toll-like receptor 4 stimulation with monophosphoryl lipid A ameliorates motor defcits and nigral neurodegeneration triggered by extraneuronal α-synucleinopathy. Mol. Neurodegener. 2017, 12, 52.
- Collins, S.M.; Surette, M.; Bercik, P. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol. 2012, 10, 735–742.
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275.
- Erny, D.; Hrabě de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977.
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139.
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358.
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015, 30, 1351–1360.
- Unger, M.M.; Spiegel, J.; Dillmann, K.U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.H. Short chain fatty acids and gut microbiota difer between patients with Parkinson’s disease and agematched controls. Parkinsonism Relat. Disord. 2016, 32, 66–67.
- Hasegawa, S.; Goto, S.; Tsuji, H.; Okuno, T.; Asahara, T.; Nomoto, K.; Shibata, A.; Fujisawa, Y.; Minato, T.; Okamoto, A.; et al. Intestinal dysbiosis and lowered serum lipopolysaccharide-binding protein in Parkinson’s disease. PLoS ONE 2015, 10, e0142164.
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 2016, 167, 1469–1480.e12.
- Cox, L.M.; Weiner, H.L. Microbiota Signaling Pathways that Influence Neurologic Disease. Neurotherapeutics 2018, 15, 135–145.
- Mayer, E.A.; Knight, R.; Mazmanian, S.K.; Cryan, J.F. Gut microbes and the brain: Paradigm shift in neuroscience. J. Neurosci. 2014, 34, 15490–15496.
- Marques, T.M.; Wall, R.; Ross, R.P.; Fitzgerald, G.F.; Ryan, C.A.; Stanton, C. Programming infant gut microbiota: Influence of dietary and environmental factors. Curr. Opin. Biotechnol. 2010, 21, 149–156.
- Cryan, J.F.; O’Mahony, S.M. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterol. Motil. 2011, 23, 187–192.
- Foster, J.A.; McVey Neufeld, K.A. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312.
- Li, P.; Killinger, B.; Ensink, E.; Beddows, I.; Yilmaz, A.; Lubben, N.; Lamp, J.; Schilthuis, M.; Vega, I.; Woltjer, R.; et al. Gut microbiota dysbiosis is associated with elevated bile acids in Parkinson’s disease. Metabolites 2021, 11, 29.
- Kang, Y.; Li, Y.; Du, Y.; Guo, L.; Chen, M.; Huang, X.; Yang, F.; Hong, J.; Kong, X. Konjaku flour reduces obesity in mice by modulating the composition of the gut microbiota. Int. J. Obesity 2019, 43, 1631–1643.
- Parashar, A.; Udayabanu, M. Gut microbiota: Implications in Parkinson’s disease. Parkinsonism Relat. Disord. 2017, 38, 1–7.
- Caputi, V.; Giron, M.C. Microbiome-gut-brain axis and toll-like receptors in Parkinson’s disease. Int. J. Mol. Sci. 2018, 19, 1689.
- Thakur, A.K.; Shakya, A.; Husain, G.M.; Emerald, M.; Kumar, V. Gut-microbiota and mental health: Current and future perspectives. J. Pharmacol. Clin. Toxicol 2014, 2, 1016.
- Forsyth, C.B.; Shannon, K.M.; Kordower, J.H.; Voigt, R.M.; Shaikh, M.; Jaglin, J.A.; Estes, J.D.; Dodiya, H.B.; Keshavarzian, A. Increased intestinal permeability correlates with sigmoid mucosa alpha-synuclein staining and endotoxin exposure markers in early Parkinson’s disease. PLoS ONE 2011, 6, e28032.
- Lin, A.; Zheng, W.; He, Y.; Tang, W.; Wei, X.; He, R.; Huang, W.; Su, Y.; Huang, Y.; Zhou, H.; et al. Gut microbiota in patients with Parkinson’s disease in southern China. Parkinsonism Relat. Disord. 2018, 53, 82–88.
- Shen, L. Gut, oral and nasal microbiota and Parkinson’s disease. Microb. Cell. Fact. 2020, 19, 50.
- Shen, X.L.; Yang, H.Z.; Wu, Y.L.; Zhang, D.F.; Jiang, H. Meta analysis: Association of helicobacter pylori infection with Parkinson’s diseases. Helicobacter 2017, 22, e12398.
- Derkinderen, P.; Shannon, K.M.; Brundin, P. Gut feelings about smoking and coffee in Parkinson’s disease. Mov. Disord. 2014, 29, 976–979.
- Li, W.; Wu, X.; Hu, X.; Wang, T.; Liang, S.; Duan, Y.; Jin, F.; Qin, B. Structural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features. Sci. China Life Sci. 2017, 60, 1223–1233.
- Hu, L.F.; Lu, M.; Tiong, C.X.; Dawe, G.S.; Hu, G.; Bian, J.S. Neuroprotective effects of hydrogen sulfide on Parkinson’s disease rat models. Aging Cell 2010, 9, 135–146.
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional implications of microbial and viral gut metagenome changes in early-stage L-DOPA-naive Parkinson’s disease patients. Genome Med. 2017, 9, 39.
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 2017, 32, 739–749.
- Qian, Y.; Yang, X.; Xu, S.; Wu, C.; Song, Y.; Qin, N.; Chen, S.D.; Xiao, Q. Alteration of the fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav. Immun. 2018, 70, 194–202.
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifrova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull. Exp. Biol. Med. 2017, 162, 734–737.
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut microbiota and metabolome alterations associated with Parkinson’s disease. mSystems 2020, 5, e00520–e00561.
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078.
- Zhao, M.; Burisch, J. Impact of genes and the environment on the pathogenesis and disease course of inflammatory bowel disease. Dig. Dis. Sci. 2019, 64, 1759–1769.
- Zhu, L.; Liu, W.; Alkhouri, R.; Baker, R.D.; Bard, J.E.; Quigley, E.M.; Baker, S.S. Structural changes in the gut microbiome of constipated patients. Physiol. Genom. 2014, 46, 679–686.
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217.
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging: An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254.
- Strasser, B.; Wolters, M.; Weyh, C.; Krüger, K.; Ticinesi, A. The Effects of Lifestyle and Diet on Gut Microbiota Composition, Inflammation and Muscle Performance in Our Aging Society. Nutrients 2021, 13, 2045.
- Kong, Y.; Wang, L.; Jiang, B. The Role of Gut Microbiota in Aging and Aging Related Neurodegenerative Disorders: Insights from Drosophila Model. Life 2021, 11, 855.
- Migliore, L.; Coppedé, F. Environmental induced oxidative stress in neurodegenerative disorders and aging. Mutat. Res. 2009, 674, 73–84.
- Tai, H.C.; Schuman, E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008, 9, 38–826.




