Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Hasan, M.; Ahn, J. Phage-Bacteria Interactions. Encyclopedia. Available online: https://encyclopedia.pub/entry/25373 (accessed on 07 February 2026).
Hasan M, Ahn J. Phage-Bacteria Interactions. Encyclopedia. Available at: https://encyclopedia.pub/entry/25373. Accessed February 07, 2026.
Hasan, Mahadi, Juhee Ahn. "Phage-Bacteria Interactions" Encyclopedia, https://encyclopedia.pub/entry/25373 (accessed February 07, 2026).
Hasan, M., & Ahn, J. (2022, July 21). Phage-Bacteria Interactions. In Encyclopedia. https://encyclopedia.pub/entry/25373
Hasan, Mahadi and Juhee Ahn. "Phage-Bacteria Interactions." Encyclopedia. Web. 21 July, 2022.
Copy Citation
With the increasing global threat of antibiotic resistance, there is an urgent need to develop new effective therapies to tackle antibiotic-resistant bacterial infections. Bacteriophage therapy is considered as a possible alternative over antibiotics to treat antibiotic-resistant bacteria. Bacteria can evolve resistance towards bacteriophages through antiphage defense mechanisms, which is a major limitation of phage therapy. The antiphage mechanisms target the phage life cycle, including adsorption, the injection of DNA, synthesis, the assembly of phage particles, and the release of progeny virions.
phage resistance
restriction modification
trade-off
antibiotic resistance
CRISPR-Cas
phage receptor
anti-phage defence system
abortive infection
superinfection exclusion
1. Introduction
Since the discovery of penicillin by Alexander Fleming in 1928, antibiotics have saved millions of lives [1]. However, at the same time, bacteria have evolved antibiotic resistance, exposing the limitation of this magic medicine to treat bacterial infections [2]. The emergence and spread of antibiotic-resistant bacteria have been accelerated due to antibiotic misuse and overuse in medicine and agriculture [3][4]. A large number of antibiotic-resistant bacteria have been identified as high-priority pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), extended-spectrum β-lactamases (ESBL) producing Enterobacteriaceae, vancomycin-resistant enterococci (VRE), and multidrug-resistant Pseudomonas aeruginosa, Streptococcus pneumonia, Mycobacterium tuberculosis, and Acinetobacter baumannii [5][6][7][8][9][10]. Human health care, animal industries, food manufacturing companies, and agricultural sectors are facing the antibiotic resistance threat [11][12]. Along with economic losses, antimicrobial resistance tolls 700,000 lives every year throughout the world due to therapeutic failure [13][14]. Bacteria can rapidly acquire molecular mechanisms to evolve antibiotic resistance through horizontal gene transfer (HGT) from everywhere [15]. The infections caused by multidrug-resistant (MDR) pathogens are difficult to treat because of the limited chemotherapeutic options, which has become a top public health concern worldwide. The frequent antibiotic treatment failures have urged the development of alternative therapeutic agents against MDR pathogens [16].
Bacteriophages (phages) are the most abundant biological entities that specifically infect bacteria [17]. Phages contribute to the diversity of bacterial communities in terms of the coevolutionary fitness dynamics [18][19]. Recently, phages have gained revived attention as alternative antibacterial agents over conventional antibiotics [20][21][22][23]. In Europe, phages have been used for therapeutic and prophylactic purposes, with a less adverse effect on normal microbial flora and no side effects [22][24]. However, the emergence of phage resistance still remains a major drawback in therapeutic applications [25][26][27]. The emergence of phage resistance is a major concern for the use of phage therapy in terms of the coevolutionary arms races between phages and bacteria [28][29]. The competitive interactions between phages and bacteria are evolutionary processes, ongoing defense strategies and counterstrategies, for survival in nature [20]. Bacteria evolve phage resistance under selection pressure, resulting in the coincidental changes in bacterial fitness and virulence [4][30].
2. Phage Structure and Life Cycle
Phages are estimated to outnumber bacteria by tenfold [31]. Phages were discovered in 1917 and first used as therapeutic agents in 1919 [32][33]. However, the interest in phage therapy was shifted to antibiotics commonly used to treat bacterial infections from the 1940s onwards. Many phages consist of a head/protein capsid containing the genomic material and tail fibers acting as receptor-binding sites [34]. Most phages have polyhedral capsids, predominantly icosahedral, except for filamentous ones [35]. The structure of tailed phages consists of an icosahedral head and a tail with receptor-binding proteins (RBPs) such as tail spikes and tail fibers at the distal end [36][37]. Phage genomes containing single-stranded or double-stranded RNA or DNA are encased in the protein capsid. RBPs are observed in Myoviridae as long and short fibers attached to the contractile tail, in Podoviridae as spikes or fibers attached to a short non-contractile tail, and in Siphoviridae as baseplates, fibers, spikes, or single straight filaments attached to a long non-contractile tail [38]. The key component of the tail is the contractile sheath. In T4 phages, the contractile sheath contracts to less than half of its original length during the infection to insert the tail tube through the outer membrane for genome delivery [39].
Phages are completely dependent on their hosts for multiplication. Phages have two distinctive life cycles, lytic and lysogenic. In the lytic life cycle, the phages infect the host, multiply inside, and lyse the host cell to release the mature progeny phages. In the lysogenic life cycle, on the contrary, phages integrate their genome into the host’s chromosome, and integrated phage DNA is replicated concurrently with bacterial DNA. In addition, certain temperate phages are maintained as episomal components rather than being incorporated into the host chromosome. The phage DNA integrated into the host cell is termed as the prophage. However, stresses such as UV radiation, antibiotics, pH, temperature, and water activity can activate prophages [32][40]. When prophage activation is triggered, an irreversible transformation from lysogeny to the lytic life cycle takes place and the phage completes the lytic cycle by making copies and lyses the cell to burst them out [41]. The phage attachment to the bacterial host commences with the binding of RBP on the tip of the phage tail to a target receptor on the host cell surface, occurring in three steps—initial contact, reversible adherence and irreversible attachment [42][43]. The initial step is the random collision of the phage with the hosts and the recognition of the receptors on the host surface [42]. After phage receptor recognition, the phage RBP reversibly binds with the receptor [44]. The last step is the irreversible phage binding to the receptor [45]. After the permanent binding with the receptor, the phage ejects the genetic material into the host cytoplasm [46] and the phage genome is expressed to produce virion particles such as head, tail, base plate, and fiber, followed by simultaneous assembly. After the phage assembly, phage-encoding enzymes such as endolysin and holin help release the progenies from the host cells [47].
3. Coevolutionary Dynamics of Phage-Bacteria Interactions
Bacteria evolve phage resistance through bacteria defense systems under phage selection pressure [48][49] (Figure 1). In this context, phages, however, evolve counter adaptations against bacterial antiphage mechanisms [27][50]. Therefore, phages and bacteria can undergo continuous coevolutionary processes involving phage infection and antiphage defense mechanisms [21][27][50][51][52]. The phage resistance mechanisms of bacteria include non-specific adaptations and specific adaptation systems [48][49]. The non-specific bacterial defense mechanisms (innate immune systems) against phages include the inhibition of phage attachment to the host surface receptors, the prevention of phage genome entry into the host cells, the restriction of secondary phage infection (superinfection exclusion), the activation of endonucleases and methyltransferases (restriction-modification system), and the induction of suicide in infected cell (abortive infection system) [48]. The phage-specific bacterial defense mechanisms (adaptive immune systems) are a second line of antiphage defense systems such as clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated (Cas) proteins [53]. The phage-resistant bacteria result in phenotypic and genotypic changes, including growth rate, membrane permeability, capsular polysaccharide (CPS) production, phage-binding receptor, virulence, and antibiotic susceptibility [54][55]. The antiphage mechanisms in bacteria developed against phage infection stages, including adsorption, penetration, synthesis, assembly, and release [56][57][58][59].
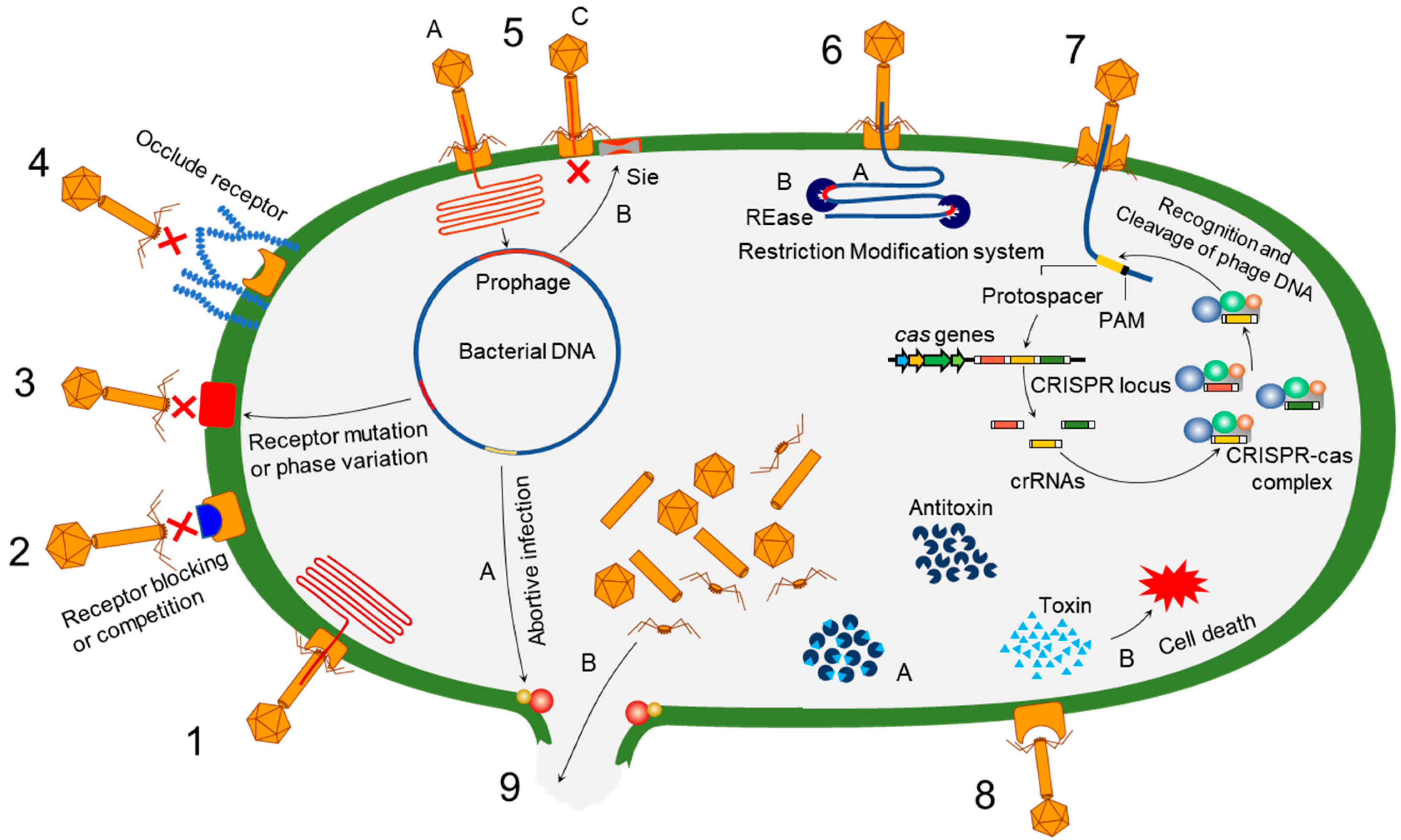
Figure 1. Overview of bacterial antiphage defense mechanisms. 1. Phage infection: successful phage attachment and infection; 2. competition in receptor binding: blocking of the phage-binding receptor to evade phage attachment; 3. alteration in cell surface receptor: mutation or phase variation in the receptor to avoid phage attachment; 4. hiding phage receptor: production of extracellular polysaccharides to hide phage receptor; 5. superinfection exclusion—A: integration of prophage into host genome; B: expression of protein to block DNA entry; C: exclusion of superinfection; 6. restriction-modification system—A: recognition of restriction site; B: cleaving of inserted phage DNA; 7. CRISPR–Cas immunity; 8. toxin–antitoxin immunity—A: antitoxin neutralizes toxin before phage infection; B: phage infection-mediated liberation of toxin to induce reduced metabolism or cell death; 9. abortive infection—A: phage infection mediates expression of abortive infection mechanism; B: release of unassembled phage particles from the host cell.
References
- Gaynes, R. The discovery of penicillin—New insights after more than 75 years of clinical use. Emerg. Infect. Dis. 2017, 23, 849–853.
- Alos, J.I. Antibiotic resistance: A global crisis. Enferm. Infec. Micribiol. Clin. 2015, 33, 692–699.
- Ojala, V.; Laitalainen, J.; Jalasvuori, M. Fight evolution with evolution: Plasmid-dependent phages with a wide host range prevent the spread of antibiotic resistance. Evol. Appl. 2013, 6, 925–932.
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216.
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–236.
- Grant, G.R.; Lederman, J.A.; Brandstetter, R.D. T.G. Heaton, tuberculosis, and artificial pneumothorax: Once again, back to the future? Chest 1997, 112, 7–8.
- Levin, A.S.; Barone, A.A.; Penco, J.; Santos, M.V.; Marinho, I.S.; Arruda, E.A.; Manrique, E.I.; Costa, S.F. Intravenous colistin as therapy for nosocomial infections caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Clin. Infect. Dis. 1999, 28, 1008–1011.
- Nachega, J.B.; Chaisson, R.E. Tuberculosis drug resistance: A global threat. Clin. Infect. Dis. 2003, 36, S24–S30.
- Rossolini, G.M.; Mantengoli, E. Antimicrobial resistance in Europe and its potential impact on empirical therapy. Clin. Microbiol. Infect. 2008, 14, 2–8.
- Whitney, C.G.; Farley, M.M.; Hadler, J.; Harrison, L.H.; Lexau, C.; Reingold, A.; Lefkowitz, L.; Cieslak, P.R.; Cetron, M.; Zell, E.R.; et al. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N. Engl. J. Med. 2000, 343, 1917–1924.
- Morehead, M.S.; Scarbrough, C. Emergence of global antibiotic resistance. Prim. Care 2018, 45, 467–484.
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. MBio 2016, 7, e02227-15.
- Pokharel, S.; Raut, S.; Adhikari, B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob. Health 2019, 4, e002104.
- Uddin, M.J.; Dawan, J.; Jeon, G.; Yu, T.; He, X.; Ahn, J. The role of bacterial membrane vesicles in the dissemination of antibiotic resistance and as promising carriers for therapeutic agent delivery. Microorganisms 2020, 8, 670.
- Wang, X.; Loh, B.; Gordillo Altamirano, F.; Yu, Y.; Hua, X.; Leptihn, S. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes Infect. 2021, 10, 2205–2219.
- Gurney, J.; Pradier, L.; Griffin, J.S.; Gougat-Barbera, C.; Chan, B.K.; Turner, P.E.; Kaltz, O.; Hochberg, M.E. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol. Med. Public Health 2020, 2020, 148–157.
- Hendrix, R.W.; Smith, M.C.M.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197.
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931.
- Hall, A.R.; Scanlan, P.D.; Morgan, A.D.; Buckling, A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol. Lett. 2011, 14, 635–642.
- Koderi Valappil, S.; Shetty, P.; Deim, Z.; Terhes, G.; Urbán, E.; Váczi, S.; Patai, R.; Polgár, T.; Pertics, B.Z.; Schneider, G.; et al. Survival comes at a cost: A coevolution of phage and its host leads to phage resistance and antibiotic sensitivity of Pseudomonas aeruginosa multidrug resistant strains. Front. Microbiol. 2021, 12, 783722.
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351.
- Hill, C.; Mills, S.; Ross, R.P. Phages and antibiotic resistance: Are the most abundant entities on earth ready for a comeback? Future Microbiol. 2018, 13, 711–726.
- Kortright, K.E.; Doss-Gollin, S.; Chan, B.K.; Turner, P.E. Evolution of bacterial cross-resistance to lytic phages and albicidin antibiotic. Front. Microbiol. 2021, 12, 658374.
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114.
- Majkowska-Skrobek, G.; Markwitz, P.; Sosnowska, E.; Lood, C.; Lavigne, R.; Drulis-Kawa, Z. The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ. Microbiol. 2021, 23, 7723–7740.
- Wright, R.C.T.; Friman, V.-P.; Smith, M.C.M.; Brockhurst, M.A. Cross-resistance is modular in bacteria-phage interactions. PLoS Biol. 2018, 16, e2006057.
- Mangalea, M.R.; Duerkop, B.A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 2020, 88, e00926-19.
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232.
- Fauconnier, A. Regulating phage therapy. EMBO Rep. 2017, 18, 198–200.
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717.
- Brussow, H.; Hendrix, R.W. Phage genomics: Small is beautiful. Cell 2002, 108, 13–16.
- Haq, I.; Chaudhry, W.; Akhtar, M.; Andleeb, S.; Qadri, I. Bacteriophages and their implications on future biotechnology: A review. Virol. J. 2012, 9, 9.
- Hermoso, J.A.; García, J.L.; García, P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007, 10, 461–472.
- Wurtz, M. Bacteriophage structure. Electron Microsc. Rev. 1992, 5, 283–309.
- Ackermann, H.W. Tailed bacteriophages: The order caudovirales. Adv. Virus Res. 1998, 51, 135–201.
- Walter, M.; Fiedler, C.; Grassl, R.; Biebl, M.; Rachel, R.; Hermo-Parrado, X.L.; Llamas-Saiz, A.L.; Seckler, R.; Miller, S.; van Raaij, M.J. Structure of the receptor-binding protein of bacteriophage det7: A podoviral tail spike in a myovirus. J. Virol. 2008, 82, 2265–2273.
- Chao, K.L.; Shang, X.; Greenfield, J.; Linden, S.B.; Alreja, A.B.; Nelson, D.C.; Herzberg, O. Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N). Sci. Rep. 2022, 12, 2061.
- Iwasaki, T.; Yamashita, E.; Nakagawa, A.; Enomoto, A.; Tomihara, M.; Takeda, S. Three-dimensional structures of bacteriophage neck subunits are shared in Podoviridae, Siphoviridae and Myoviridae. Genes Cells 2018, 23, 528–536.
- Aksyuk, A.A.; Leiman, P.G.; Kurochkina, L.P.; Shneider, M.M.; Kostyuchenko, V.A.; Mesyanzhinov, V.V.; Rossmann, M.G. The tail sheath structure of bacteriophage T4: A molecular machine for infecting bacteria. EMBO J. 2009, 28, 821–829.
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251.
- Xu, J.; Kiesel, B.; Kallies, R.; Jiang, F.L.; Liu, Y.; Maskow, T. A fast and reliable method for monitoring of prophage-activating chemicals. Microb. Biotechnol. 2018, 11, 1112–1120.
- Moldovan, R.G.; Chapman-McQuiston, E.; Wu, X.L. On kinetics of phage adsorption. Biophys. J. 2007, 93, 303–315.
- Quiberoni, A.; Guglielmotti, D.; Binetti, A.; Reinheimer, J. Characterization of three Lactobacillus delbrueckii subsp. bulgaricus phages and the physicochemical analysis of phage adsorption. J. Appl. Microbiol. 2004, 96, 340–351.
- Uchiyama, J.; Takemura, I.; Satoh, M.; Kato, S.; Ujihara, T.; Akechi, K.; Matsuzaki, S.; Daibata, M. Improved adsorption of an Enterococcus faecalis bacteriophage PhiEF24C with a spontaneous point mutation. PLoS ONE 2011, 6, e26648.
- Dowah, A.S.A.; Clokie, M.R.J. Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 2018, 10, 535–542.
- Egido, J.E.; Costa, A.R.; Aparicio-Maldonado, C.; Haas, P.J.; Brouns, S.J.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. 2022, 46, fuab048.
- Kasman, L.M.; Porter, L.D. Bacteriophages. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022.
- Bikard, D.; Marraffini, L.A. Innate and adaptive immunity in bacteria: Mechanisms of programmed genetic variation to fight bacteriophages. Curr. Opin. Immunol. 2012, 24, 15–20.
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687.
- Burmeister, A.R.; Sullivan, R.M.; Gallie, J.; Lenski, R.E. Sustained coevolution of phage Lambda and Escherichia coli involves inner- as well as outer-membrane defences and counter-defences. Microbiology 2021, 167, 1063.
- Scanlan, P.D.; Buckling, A. Co-evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J. 2012, 6, 1148–1158.
- Scanlan, P.D.; Hall, A.R.; Lopez-Pascua, L.D.; Buckling, A. Genetic basis of infectivity evolution in a bacteriophage. Mol. Ecol. 2011, 20, 981–989.
- Isaev, A.B.; Musharova, O.S.; Severinov, K.V. Microbial arsenal of antiviral defenses—Part I. Biochemistry 2021, 86, 319–337.
- Kortright, K.E.; Done, R.E.; Chan, B.K.; Souza, V.; Turner, P.E.; Vives, M. Selection for phage resistance reduces virulence of Shigella flexneri. Appl. Environ. Microbiol. 2022, 88, e01514-21.
- Capparelli, R.; Nocerino, N.; Lanzetta, R.; Silipo, A.; Amoresano, A.; Giangrande, C.; Becker, K.; Blaiotta, G.; Evidente, A.; Cimmino, A.; et al. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS ONE 2010, 5, e11720.
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134.
- Summers, W.C. The strange history of phage therapy. Bacteriophage 2012, 2, 130–133.
- Hampton, H.G.; Watson, B.N.J.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336.
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
2 times
(View History)
Update Date:
21 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




