Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Megan I Mitchell | -- | 2488 | 2022-07-19 15:40:54 | | | |
| 2 | Conner Chen | + 2 word(s) | 2490 | 2022-07-20 08:53:00 | | | | |
| 3 | Conner Chen | + 2 word(s) | 2492 | 2022-07-20 08:55:15 | | | | |
| 4 | Conner Chen | -5 word(s) | 2487 | 2022-07-20 09:59:01 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Mitchell, M.I.; Ma, J.; Carter, C.L.; Loudig, O. Exosome Isolation Techniques. Encyclopedia. Available online: https://encyclopedia.pub/entry/25285 (accessed on 07 February 2026).
Mitchell MI, Ma J, Carter CL, Loudig O. Exosome Isolation Techniques. Encyclopedia. Available at: https://encyclopedia.pub/entry/25285. Accessed February 07, 2026.
Mitchell, Megan I., Junfeng Ma, Claire L. Carter, Olivier Loudig. "Exosome Isolation Techniques" Encyclopedia, https://encyclopedia.pub/entry/25285 (accessed February 07, 2026).
Mitchell, M.I., Ma, J., Carter, C.L., & Loudig, O. (2022, July 19). Exosome Isolation Techniques. In Encyclopedia. https://encyclopedia.pub/entry/25285
Mitchell, Megan I., et al. "Exosome Isolation Techniques." Encyclopedia. Web. 19 July, 2022.
Copy Citation
Extracellular vesicles (EVs) are lipid-binding vesicles secreted by cells into the extracellular space. The technologies and tools that have been used to purify exosomes from blood and other biofluids have evolved greatly, and this evolution has been driven by the need to accurately assess their biological function, but also and most importantly to decipher their molecular content, with a particular focus on tumor exosome biomarkers
Extracellular Vesicles
Exosomes
Circulating Biomarkers
Isolation Techniques
1. Ultracentrifugation-Based Isolation
Exosome isolation by means of ultracentrifugation is still the most widely used approach and is considered the gold standard for sedimentation of exosomes [1][2]. To date, it is estimated that ultracentrifugation accounts for ~56% of all exosome isolation techniques, as it is demonstrated to provide high-purity exosome fractions from biofluids [3]. Currently, two protocols for ultracentrifugation-based exosome isolations are used, including either differential ultracentrifugation or density gradient ultracentrifugation [4].
Differential ultracentrifugation: consists of a series of sequential centrifugation cycles at different centrifugal forces and durations [4]. The initial preparation of biofluids typically starts with several short, low-speed centrifugation steps (i.e., 2000× g for 10 min followed by 10,000 × g for 30 min [1]) that are necessary for the removal of contaminating cellular debris and larger microvesicles [5]. Next, a first round of ultracentrifugation is carried out at ~100,000× g for 90 min to generate an exosome pellet, which is washed with an appropriate isotonic buffer such as phosphate buffered saline (PBS) to remove protein and other soluble molecules. Subsequently, a second ultracentrifugation round (i.e., at ~100,000× g for 90 min) is performed to obtain the final exosome pellet, which is generally re-suspended in PBS and stored at −80 °C to await downstream analyses. Studies have also shown that exosomes purified by ultracentrifugation can be stably stored at 4 °C, where they maintain their intactness and retain their function for up to 20 months [6].
Density-gradient ultracentrifugation: (standard or isopycnic) has recently gained popularity because studies have shown that it increases the purity of exosome preparations [7]. Here, the separation of exosomes is achieved by the layering of a liquid sample as a narrow band on top of a medium, typically sucrose or iodixanol [8]. With the application of centrifugal force (i.e., at ~100,000× g for up to 18 h), the gradient allows for the separation of solutes, including exosomes, and their specific sedimentation into several distinct solute layers. After centrifugation, individual 1 mL gradient fractions are manually collected using a pipette [9]. The separated exosome fraction is then diluted with 1x PBS and subjected to a second round of ultracentrifugation (i.e., at ~100,000× g for ~70 min [7]). Similar to standard ultracentrifugation, the resulting exosome pellet is resuspended in PBS and stored at −80 °C. The biggest limitation when selecting density-gradient ultracentrifugation over differential ultracentrifugation is that the sample volume capacity for exosome isolation is greatly reduced in the former (~5% of the centrifuge tube capacity) [10].
Although ultracentrifugation remains the gold-standard for sedimentation of exosomes devoid of other extracellular vesicles (EVs) (i.e., larger sized microvesicles, cell debris, proteins) and lipoprotein contaminants, it requires expensive instrumentation, but most importantly only provides a bulk exosome isolate from a specific biofluid, rather that separating cell/tissue-specific exosome sub-populations from a biofluid [11]. Additionally, recent studies have reported that repeated rounds of ultracentrifugation reduce exosome yield and that extended and repeated high shear forces from high-speed ultracentrifugation adversely affect their integrity [12][13].
2. Size-Based Separation and Isolation
To date there are three main methods for the size-based isolation of exosomes, which include ultrafiltration, sequential filtration, and size-exclusion chromatography (SEC). These methods have been developed to bypass extended ultracentrifugation times and to facilitate the fractionation of exosomes from large biofluid volumes.
Ultrafiltration: relies on the use of 10–100 kDa molecular weight cutoff (MWCO) filters, which are most often utilized to reduce large sample volumes (i.e., urine or cell culture supernatants) by concentration to smaller more manageable volumes that often simplify downstream applications [14]. Currently, several commercially available kits are on the market which have been proven to provide pure populations of exosomes from biofluids. These include the Qiagen exoEasy kit, which allows isolation of exosomes from volumes up to 4 mL of plasma or 16 mL of cell culture media [15]; the Amicon® Ultra-15 centrifugal filter tubes (MilliporeSigma, Burlington, MA, USA), which allow filtration of up to 15 mL [16]; and the ExoLution® plus platform from Exosome Diagnostics, which allows the processing of 3 mL of plasma [17].
Sequential filtration:, unlike standard ultrafiltration, typically includes three steps [18]. First, larger cells and cellular debris are filtered out using standard filtration methods (0.2 µm filters). Second, free proteins are depleted by tangential flow filtration, where biofluids are passed parallel to the filter, allowing for continuous filtration and reduced membrane clogging [19]. Third, exosomes are sorted out with the use of a track-etched filter membrane (e.g., Whatman/GE Nucleopore 50, 80, 100, or 200 nm diameter membranes), allowing for size-based fractionation of exosomes [20].
Size exclusion chromatography (SEC): relies on passing a biofluid (known as the mobile phase) through a porous gel filtration polymer (i.e., the stationary phase [21]). The stationary phase of the chromatography column can be packed with different types of polymers (e.g., agarose, polyacrylamide, crosslinked dextrans, or allyldextran), the nature of which allows for differential elution of the sample into size-based fractions (i.e., larger particles travel faster and elute first, followed by smaller particles and finally non-membrane bound proteins [22]). In terms of its use for exosome isolation, SEC, when performed at lower flow rates, has been shown to possess several advantages, including reproducibility, cost efficiency, and the isolation of undamaged exosomes [12]. SEC significantly outcompetes other filtration-based isolation techniques in that it effectively removes protein contaminants, thereby yielding highly pure exosome fractions [23]. The isolation, purification, and enrichment of exosomes using SEC has been successfully used with a variety of biological fluids, including serum and plasma [24], cerebrospinal fluid [25], bovine and human milk [26], saliva [27], urine [28], synovial fluid [29], tears [27], nasal lavage [30], and seminal fluid [31].
Although great purity is achieved via SEC, which can accommodate large volumes of sample (i.e., up to 100 mL using the qEV100 column from IZON), this approach does not allow for separation of different exosome subpopulations or for separation of exosomes from other vesicles of the same size; thus, contamination by lipoproteins and other microvesicles cannot be ruled out [32].
3. Polyethylene Glycol (PEG) Precipitation-Based Isolation
The isolation of exosomes can also be achieved by precipitation by the addition of an aqueous polyethylene glycol (PEG) solution to a biofluid. Here, PEG coats the surface of exosomes and other microvesicles, facilitating the formation of exosome–PEG aggregates [33]. Exosomes are trapped in this porous microstructure, and are then precipitated (e.g., ExoQuick®) by low-speed centrifugation at 1500× g [34]. This method results in the isolation of exosomes within a size range consistent with ultracentrifugation; however, as PEG is a non-specific “coagulant”, it also results in the co-precipitation of soluble non-exosomal proteins, immunoglobulins, and lipoproteins, which significantly limits the purity of the final exosome pellet [35] as they carry unwanted biological material (i.e., proteins and nucleic acid species) [36]. The advantages of this procedure are that it is inexpensive, requires little to no training, and it allows for high-throughput processing of samples with little damage to exosomes. Although PEG-based precipitation results in high yield, low purity exosomes, studies have shown that when it is sequentially combined with an immuno-purification (such as anti-CD63 assay), the exosome fraction purity may be enhanced [37], making it an attractive approach for initial, fast, and crude isolation of exosomes [38].
4. Immunoaffinity-Based Isolation
In recent years, immunoaffinity-based capture has become one of the preferred methods for the isolation of exosomes from biofluids [39]. This technology allows for direct separation of exosomes via immobilized antibody targeting of membrane-surface specific exosomal proteins [40][41][42]. Most, if not all, commercially available immunoaffinity-based isolation kits are tailored with antibodies (either alone or in combination, i.e., a pan-exosome panel) targeting common exosome surface markers, such as the tetraspanins (CD9, CD63, and CD81) [43]. Other assays also include antibodies targeting markers such as epithelial cellular adhesion molecule (EPCAM) [44] and exosome-binding molecules such as heat shock proteins (HSP70, HSP90) [45] or heparin [46]. A large number of biotechnology companies, including MBL (EXOCAP, Sunnyvale, CA, USA), Systems Bioscience (ExoFlow, Palo Alto, Santa Clara, CA, USA), FujiFilm (MagCapture™, Wako Chemicals, Richmond, VA, USA), and BioLegend (MojoSort™, San Diego, CA, USA), provide ready-to-use immunoaffinity assays for exosome isolation [36] that utilize antibody-coated magnetic beads for the capture of exosomes, as they are convenient, allow rapid and easy magnetic isolation of exosomes, and ultimately allow for their concentration into a small volume [47]. Although the selection of exosomes from a biofluid, with antibodies targeting exosome surface proteins, provides the state-of-the-art selection, very few provide assays that allow for the gentle release of captured exosomes [40]. Another critical issue with magnetic beads, as determined by quantitative PCR of small-RNA contaminants, is that they can bind small-RNAs and EVs (containing small-RNAs), which may be detrimental for downstream analyses, especially when analyzing the small-RNA content of exosome sub-populations that are in low abundance [40]. It can be hypothesized that the polymer coating is positively charged and the pore sizes between adjacent streptavidin/carboxyl molecules may be large enough to allow for non-specific binding of small biological material (i.e., nucleic acids, proteins, and lipids), creating a non-specific signal background that will interfere with subsequent analyses and the identification of exosome tumor biomarkers (i.e., transcriptomic signatures). To circumvent these limitations, a group recently developed an assay that was termed EV-CATCHER (Extracellular Vesicles Capture by AnTibody of CHoice and Enzymatic Release), which greatly reduces non-specific binding of small-RNAs and EVs, which can be customized with any antibody, and which allows for the mild enzymatic release of intact EVs [40].
Overall, immunoaffinity-based capture generally reduces exosome yield (as only antibody-recognized exosomes are captured) and is generally more expensive; however, when coupled with an initial step of ultracentrifugation or ultrafiltration to concentrate exosomes, it can result in higher exosome purity and may help separate exosomes of specific cellular origin from the bulk exosome preparation. Many studies on lung [48], breast [49], colorectal [50], liver [51], and many other cancers have already demonstrated that with higher exosome purity and selectivity, the analysis of their molecular cargo can provide more specific and reproducibly detectable biomarkers [52][53]. Thus, the immuno-affinity separation of unique tumor exosome cargoes from biofluids has potential to lead to novel cancer diagnostic assays [54].
5. Microfluidics-Based Isolation
Microfluidics-based exosome isolation systems have become a sought-after nanotechnology for separating exosomes from other nanosized bioparticles. This technology provides high-speed, high-throughput, ultra-precise, and cost-efficient isolation of exosomes [55]. Emerging chip-based microfluidic exosome isolation techniques, including standard research laboratory-based approaches developed by Duke University using acoustic microfluidics (i.e., acoustofluidics) for the separation of exosomes from whole blood [56], the exosome total isolation chip (ExoTIC) device developed by Liu et al., and the ExoChip developed by Kanwar et al., have already been marketed for efficient isolation of high-yield, high-purity intact exosomes [57][58]. The ExoTIC device utilizes a simple filtration-based approach wherein EV-containing biofluids, such as culture media, plasma, and urine, are passed through a nanoporous membrane [57]. During this process, free-floating proteins, nucleic acids, and lipids are washed out, and the enriched exosomes (30–200 nm in size) can be collected from the membrane using a standard pipette [57]. In comparison to ultracentrifugation and precipitation-based exosome isolation, the ExoTIC device has been shown to possess undisputed advantages, which include the constant flow of clinical specimens through nanochannels that slow the EVs, the separation of large and small vesicles in separate nano-sized chambers via engineered ports, and the presence of antibodies for the capture of exosomes with potential for release [59]. Currently, many of the existing microfluidics-based systems, including acoustic microfluidics, ExoTIC, and ExoChip, incorporate immunoaffinity components for the separation and capture of exosomes by targeting specific surface markers with immobilized antibodies [58][60]. Interestingly, although a recent analyses revealed the strong limitations of magnetic beads for isolating pure exosomes, several microfluidic platforms have incorporated magnetic beads into their systems for the separation of cells from biofluids [57][58]. Currently, some of the most widely used microfluidics-based systems fully integrate size-based separation, immunoaffinity-based separation, and dynamic separation [61]. The most critical issue that microfluidic-based separation technologies have solved is that they avoid the non-continuous separation processes. Indeed, samples are processed from start-to-finish in a single run, in a closed loop, with limited user-interference or introduction of contaminants, which helps maintain yields that are otherwise reduced by the repeated washes performed during sequential ultracentrifugation [12]. Several problems still remain, however, which are similar to those mentioned for immunoaffinity-based isolation techniques and include the need for high immunoaffinity and the requirement for highly specific and sensitive antibodies [62]. Additionally, while microfluidic systems require less hands-on manipulation, they require expensive equipment and complex nano-sized chips that are often difficult to mass-produce [63]. Despite these challenges that remain to be solved, microfluidics-based purifications offer a promising technology for all-in-one chip-based robust isolation and characterization of circulating tumor exosome biomarkers.
6. Exosome Sorting by Fluorescence Activated Cell Sorting (FACS)
The separation of cells by means of flow cytometry is commonplace [64][65][66]. However, because exosomes are 30–150 nm in size, they fall under the threshold of even the most sensitive “reference noise” region of flow cytometers [67][68]. Nevertheless, selective sorting of antibody-labeled exosome sub-populations by FACS shows great potential for cancer biomarker screening and discovery. As such, extensive research efforts have been applied to the development of high-fidelity-based FACS systems for sorting exosomes [69][70][71] for subsequent proteomic and transcriptomic analyses [72]. One of the biggest advancements has been the development and marketing of advanced imaging flow cytometry (iFCM) by ImageStreamx (ISx, EMD Millipore, Seattle, WA, USA) [73] which offers significant advantages for the sorting of exosomes and other small EVs when compared to other available technologies [73][74]. Mastoridis et al. demonstrated that by combining iFCM with subset-specific markers, this technology allows for the high-throughput, multiparametric characterization, and functional assessment of exosomes [75]. Another promising avenue for FACS-based sorting and cancer diagnostics based on the presence of unique tumor exosomes in a biofluid (i.e., prostate cancer exosomes) is the multicolor multiplexed in situ proximity ligation, also termed exoPLA, developed in the laboratory of Dr. Kamali-Moghaddam [76][77]. This method relies on four oligonucleotide-conjugated antibodies whose combination uniquely targets a specific type of exosomes (i.e., prostate exosomes) and rolling circle PCR amplification guided by individual pairs of antibodies to generate different fluorescent signals. Ultimately, the combination of three different fluorescent signals allows for the unique detection and sorting of tumor exosomes by flow cytometry [78]. Altogether, while the FACS-based sorting of exosomes is still a relatively new method in comparison to the others described, it provides a promising avenue for highly specific and robust isolation of low-abundance exosome sub-populations and subsequent molecular analyses.
Collectively, while most of the described exosome isolation techniques are currently available and used in different applications (Figure 1), they all possess advantages and disadvantages, which need to be considered prior to initiating exosome analyses. The ideal method for exosome isolation should be relatively simple, efficient, inexpensive, and scalable, but most importantly ultra-sensitive in order to allow for the robust and accurate identification of encapsulated circulating tumor biomarkers and to propel exosome-based liquid biopsies toward ultra-sensitive cancer diagnostics.
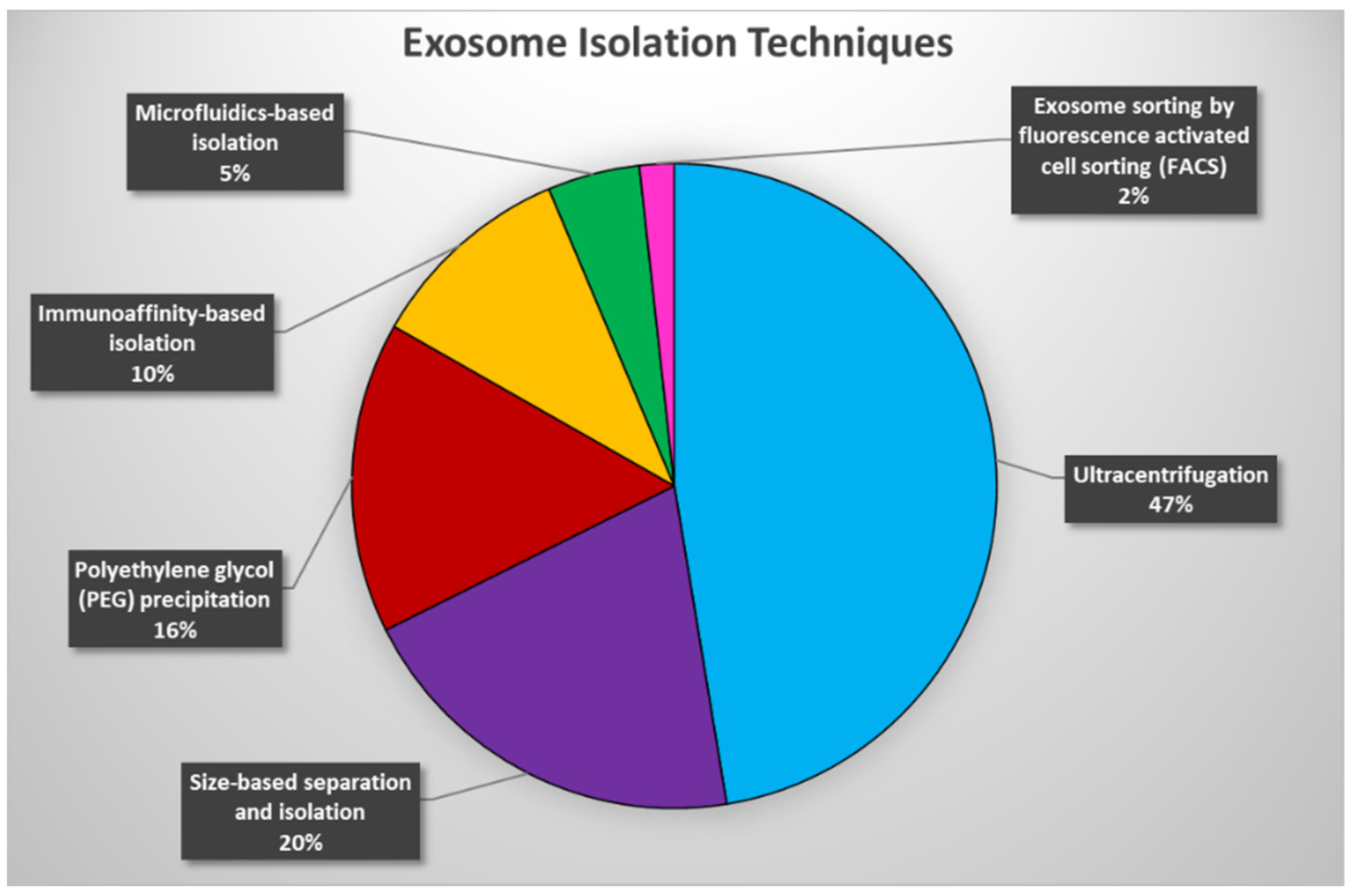
Figure 1. An estimate of the use of exosome isolation techniques over the past 10 years. Pie chart representation of the percentage utilization of each exosome isolation technique extracted from 173 publications spanning the years 2012–2022.
References
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031.
- Liangsupree, T.; Multia, E.; Riekkola, M.-L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2020, 1636, 461773.
- Li, W.J.; Chen, H.; Tong, M.L.; Niu, J.J.; Zhu, X.Z.; Lin, L.R. Comparison of the yield and purity of plasma exosomes extracted by ultracentrifugation, precipitation, and membrane-based approaches. Open Chem. 2022, 20, 182–191.
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319, Erratum in Sci. Rep. 2016, 6, 21447. Livshts, Mikhail A .
- Purushothaman, A. Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. Methods Mol. Biol. 2019, 1952, 233–244.
- Kumeda, N.; Ogawa, Y.; Akimoto, Y.; Kawakami, H.; Tsujimoto, M.; Yanoshita, R. Characterization of Membrane Integrity and Morphological Stability of Human Salivary Exosomes. Biol. Pharm. Bull. 2017, 40, 1183–1191.
- Zhang, Z.; Wang, C.; Li, T.; Liu, Z.; Li, L. Comparison of ultracentrifugation and density gradient separation methods for isolating Tca8113 human tongue cancer cell line-derived exosomes. Oncol. Lett. 2014, 8, 1701–1706.
- Brakke, M.K. Density gradient centrifugation. A new centrifugation technique. J. Am. Chem. Soc. 1951, 73, 1847.
- Dhondt, B.; Lumen, N.; De Wever, O.; Hendrix, A. Preparation of Multi-omics Grade Extracellular Vesicles by Density-Based Fractionation of Urine. STAR Protoc. 2020, 1, 100073.
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707.
- Ayala-Mar, S.; Donoso-Quezada, J.; Gallo-Villanueva, R.C.; Perez-Gonzalez, V.H.; González-Valdez, J. Recent advances and challenges in the recovery and purification of cellular exosomes. Electrophoresis 2019, 40, 3036–3049.
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.G.; Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017, 13, 2061–2065.
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 8545347.
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol. Ther. 2018, 26, 2838–2847.
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1.
- Wang, W.; Lian, J.Q.; Wang, P.Z.; Pan, L.; Ji, X.Y.; Bai, X.F.; Jia, Z.S. Isolation of exosomes derived from dendritic cells by ultrafiltration centrifugalization and their morphologic characteristics. J. Cell. Mol. Immunol. 2007, 23, 1119–1121. (In Chinese)
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.G.; Castellanos-Rizaldos, E.; et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018, 29, 700–706.
- Heinemann, M.L.; Ilmer, M.; Silva, L.P.; Hawke, D.H.; Recio, A.; Vorontsova, M.A.; Alt, E.; Vykoukal, J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A 2014, 1371, 125–135.
- van Reis, R.; Gadam, S.; Frautschy, L.N.; Orlando, S.; Goodrich, E.M.; Saksena, S.; Kuriyel, R.; Simpson, C.M.; Pearl, S.; Zydney, A.L. High performance tangential flow filtration. Biotechnol. Bioeng. 1997, 56, 71–82.
- Xu, R.; Greening, D.W.; Zhu, H.J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162.
- An, M.; Wu, J.; Zhu, J.; Lubman, D.M. Comparison of an Optimized Ultracentrifugation Method versus Size-Exclusion Chromatography for Isolation of Exosomes from Human Serum. J. Proteome Res. 2018, 17, 3599–3605.
- Gaspar, L.S.; Santana, M.M.; Henriques, C.; Pinto, M.M.; Ribeiro-Rodrigues, T.M.; Girão, H.; Nobre, R.J.; Pereira de Almeida, L. Simple and Fast SEC-Based Protocol to Isolate Human Plasma-Derived Extracellular Vesicles for Transcriptional Research. Mol. Ther. Methods Clin. Dev. 2020, 18, 723–737.
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466.
- Wei, R.; Zhao, L.; Kong, G.; Liu, X.; Zhu, S.; Zhang, S.; Min, L. Combination of Size-Exclusion Chromatography and Ultracentrifugation Improves the Proteomic Profiling of Plasma-Derived Small Extracellular Vesicles. Biol. Proced. Online 2020, 22, 12.
- Welton, J.L.; Loveless, S.; Stone, T.; von Ruhland, C.; Robertson, N.P.; Clayton, A. Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis. J. Extracell. Vesicles 2017, 6, 1369805.
- Vaswani, K.; Mitchell, M.D.; Holland, O.J.; Qin Koh, Y.; Hill, R.J.; Harb, T.; Davies, P.S.W.; Peiris, H. A Method for the Isolation of Exosomes from Human and Bovine Milk. J. Nutr. Metab. 2019, 2019, 5764740.
- Aqrawi, L.A.; Galtung, H.K.; Vestad, B.; Øvstebø, R.; Thiede, B.; Rusthen, S.; Young, A.; Guerreiro, E.M.; Utheim, T.P.; Chen, X.; et al. Identification of potential saliva and tear biomarkers in primary Sjögren’s syndrome, utilising the extraction of extracellular vesicles and proteomics analysis. Arthritis Res. Ther. 2017, 19, 14.
- Oeyen, E.; Van Mol, K.; Baggerman, G.; Willems, H.; Boonen, K.; Rolfo, C.; Pauwels, P.; Jacobs, A.; Schildermans, K.; Cho, W.C.; et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J. Extracell. Vesicles 2018, 7, 1490143.
- Foers, A.D.; Chatfield, S.; Dagley, L.F.; Scicluna, B.J.; Webb, A.I.; Cheng, L.; Hill, A.F.; Wicks, I.P.; Pang, K.C. Enrichment of extracellular vesicles from human synovial fluid using size exclusion chromatography. J. Extracell. Vesicles 2018, 7, 1490145.
- Bartel, S.; La Grutta, S.; Cilluffo, G.; Perconti, G.; Bongiovanni, A.; Giallongo, A.; Behrends, J.; Kruppa, J.; Hermann, S.; Chiang, D.; et al. Human airway epithelial extracellular vesicle miRNA signature is altered upon asthma development. Allergy 2020, 75, 346–356.
- Rodriguez-Caro, H.; Dragovic, R.; Shen, M.; Dombi, E.; Mounce, G.; Field, K.; Meadows, J.; Turner, K.; Lunn, D.; Child, T.; et al. In vitro decidualisation of human endometrial stromal cells is enhanced by seminal fluid extracellular vesicles. J. Extracell. Vesicles 2019, 8, 1565262.
- Takov, K.; Yellon, D.M.; Davidson, S.M. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: Yield, purity and functional potential. J. Extracell. Vesicles 2018, 8, 1560809.
- Buschmann, D.; Mussack, V.; Byrd, J.B. Separation, characterization, and standardization of extracellular vesicles for drug delivery applications. Adv. Drug Deliv. Rev. 2021, 174, 348–368.
- Fang, X.; Duan, Y.; Adkins, G.B.; Pan, S.; Wang, H.; Liu, Y.; Zhong, W. Highly Efficient Exosome Isolation and Protein Analysis by an Integrated Nanomaterial-Based Platform. Anal. Chem. 2018, 90, 2787–2795.
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646.
- Hoffmann, M.M.; Bothe, S.; Gutmann, T.; Buntkowsky, G. Unusual local molecular motions in the solid state detected by dynamic nuclear polarization enhanced NMR spectroscopy. J. Phys. Chem. C 2017, 121, 22948–22957.
- Shu, S.; Yang, Y.; Allen, C.L.; Hurley, E.; Tung, K.H.; Minderman, H.; Wu, Y.; Ernstoff, M.S. Purity and yield of melanoma exosomes are dependent on isolation method. J. Extracell. Vesicles 2019, 9, 1692401.
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208.
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2022, 9, 811971.
- Mitchell, M.I.; Ben-Dov, I.Z.; Liu, C.; Ye, K.; Chow, K.; Kramer, Y.; Gangadharan, A.; Park, S.; Fitzgerald, S.; Ramnauth, A.; et al. Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): A customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs. J. Extracell. Vesicles 2021, 10, e12110.
- Wu, D.; Yan, J.; Shen, X.; Sun, Y.; Thulin, M.; Cai, Y.; Wik, L.; Shen, Q.; Oelrich, J.; Qian, X.; et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun. 2019, 10, 3854.
- Hu, Q.; Su, H.; Li, J.; Lyon, C.; Tang, W.; Wan, M.; Hu, T.Y. Clinical applications of exosome membrane proteins. Precis. Clin. Med. 2020, 3, 54–66.
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568.
- Zhou, Y.G.; Mohamadi, R.M.; Poudineh, M.; Kermanshah, L.; Ahmed, S.; Safaei, T.S.; Stojcic, J.; Nam, R.K.; Sargent, E.H.; Kelley, S.O. Interrogating Circulating Microsomes and Exosomes Using Metal Nanoparticles. Small 2016, 12, 727–732.
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE 2014, 9, e110443.
- Balaj, L.; Atai, N.A.; Chen, W.; Mu, D.; Tannous, B.A.; Breakefield, X.O.; Skog, J.; Maguire, C.A. Heparin affinity purification of extracellular vesicles. Sci. Rep. 2015, 5, 10266.
- Koliha, N.; Wiencek, Y.; Heider, U.; Jüngst, C.; Kladt, N.; Krauthäuser, S.; Johnston, I.C.; Bosio, A.; Schauss, A.; Wild, S. A novel multiplex bead-based platform highlights the diversity of extracellular vesicles. J. Extracell. Vesicles 2016, 5, 29975.
- Nie, H.; Xie, X.; Zhang, D.; Zhou, Y.; Li, B.; Li, F.; Li, F.; Cheng, Y.; Mei, H.; Meng, H.; et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale 2020, 12, 877–887.
- Gotanda, K.; Hirota, T.; Saito, J.; Fukae, M.; Egashira, Y.; Izumi, N.; Deguchi, M.; Kimura, M.; Matsuki, S.; Irie, S.; et al. Circulating intestine-derived exosomal miR-328 in plasma, a possible biomarker for estimating BCRP function in the human intestines. Sci. Rep. 2016, 6, 32299.
- Sun, H.; Meng, Q.; Shi, C.; Yang, H.; Li, X.; Wu, S.; Familiari, G.; Relucenti, M.; Aschner, M.; Wang, X.; et al. Hypoxia-Inducible Exosomes Facilitate Liver-Tropic Premetastatic Niche in Colorectal Cancer. Hepatology 2021, 74, 2633–2651.
- Xu, Y.F.; Hannafon, B.N.; Zhao, Y.D.; Postier, R.G.; Ding, W.Q. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget 2017, 8, 77028–77040.
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465.
- Padda, J.; Khalid, K.; Khedr, A.; Patel, V.; Al-Ewaidat, O.A.; Tasnim, F.; Padda, S.; Cooper, A.C.; Jean-Charles, G. Exosome-Derived microRNA: Efficacy in Cancer. Cureus 2021, 13, e17441.
- Lou, D.; Wang, Y.; Yang, Q.; Hu, L.; Zhu, Q. Ultrafiltration combing with phospholipid affinity-based isolation for metabolomic profiling of urinary extracellular vesicles. J. Chromatogr. A 2021, 1640, 461942.
- Salafi, T.; Zeming, K.K.; Zhang, Y. Advancements in microfluidics for nanoparticle separation. Lab. Chip. 2016, 17, 11–33.
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589, Erratum in Proc. Natl. Acad. Sci. USA 2020, 117, 28525.
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723.
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab. Chip. 2014, 14, 1891–1900.
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small 2020, 16, e1903916.
- Gou, Y.; Jia, Y.; Wang, P.; Sun, C. Progress of Inertial Microfluidics in Principle and Application. Sensors 2018, 18, 1762.
- Ding, L.; Yang, X.; Gao, Z.; Effah, C.Y.; Zhang, X.; Wu, Y.; Qu, L. A Holistic Review of the State-of-the-Art Microfluidics for Exosome Separation: An Overview of the Current Status, Existing Obstacles, and Future Outlook. Small 2021, 17, e2007174.
- Yang, F.; Liao, X.; Tian, Y.; Li, G. Exosome separation using microfluidic systems: Size-based, immunoaffinity-based and dynamic methodologies. Biotechnol. J. 2017, 12, 1600699.
- Lee, H.; Lee, J.; Lee, S.G.; Doyle, P.S. Hydrogel-Based Colorimetric Assay for Multiplexed MicroRNA Detection in a Microfluidic Device. Anal. Chem. 2020, 92, 5750–5755.
- Liao, X.; Makris, M.; Luo, X.M. Fluorescence-activated Cell Sorting for Purification of Plasmacytoid Dendritic Cells from the Mouse Bone Marrow. J. Vis. Exp. 2016, 117, e54641.
- Kindlund, B.; Sjöling, Å.; Yakkala, C.; Adamsson, J.; Janzon, A.; Hansson, L.E.; Hermansson, M.; Janson, P.; Winqvist, O.; Lundin, S.B. CD4+ regulatory T cells in gastric cancer mucosa are proliferating and express high levels of IL-10 but little TGF-β. Gastric Cancer 2017, 20, 116–125.
- Malmberg, E.B.; Ståhlman, S.; Rehammar, A.; Samuelsson, T.; Alm, S.J.; Kristiansson, E.; Abrahamsson, J.; Garelius, H.; Pettersson, L.; Ehinger, M.; et al. Patient-tailored analysis of minimal residual disease in acute myeloid leukemia using next-generation sequencing. Eur. J. Haematol. 2017, 98, 26–37.
- Richards, A.J.; Staats, J.; Enzor, J.; McKinnon, K.; Frelinger, J.; Denny, T.N.; Weinhold, K.J.; Chan, C. Setting objective thresholds for rare event detection in flow cytometry. J. Immunol. Methods 2014, 409, 54–61.
- Morales-Kastresana, A.; Musich, T.A.; Welsh, J.A.; Telford, W.; Demberg, T.; Wood, J.C.S.; Bigos, M.; Ross, C.D.; Kachynski, A.; Dean, A.; et al. High-fidelity detection and sorting of nanoscale vesicles in viral disease and cancer. J. Extracell. Vesicles 2019, 19, 1597603.
- Cao, Z.; Li, C.; Higginbotham, J.N.; Franklin, J.L.; Tabb, D.L.; Graves-Deal, R.; Hill, S.; Cheek, K.; Jerome, W.G.; Lapierre, L.A.; et al. Use of fluorescence-activated vesicle sorting for isolation of Naked2-associated, basolaterally targeted exocytic vesicles for proteomics analysis. Mol. Cell Proteom. 2008, 7, 1651–1667.
- Poncelet, P.; Robert, S.; Bouriche, T.; Bez, J.; Lacroix, R.; Dignat-George, F. Standardized counting of circulating platelet microparticles using currently available flow cytometers and scatter-based triggering: Forward or side scatter? Cytom. A 2016, 89, 148–158.
- Chandler, W.L.; Yeung, W.; Tait, J.F. A new microparticle size calibration standard for use in measuring smaller microparticles using a new flow cytometer. J. Thromb. Haemost. 2011, 9, 1216–1224.
- Groot Kormelink, T.; Arkesteijn, G.J.; Nauwelaers, F.A.; van den Engh, G.; Nolte-‘t Hoen, E.N.; Wauben, M.H. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytom. A 2016, 89, 135–147.
- Padda, R.S.; Deng, F.K.; Brett, S.I.; Biggs, C.N.; Durfee, P.N.; Brinker, C.J.; Williams, K.C.; Leong, H.S. Nanoscale flow cytometry to distinguish subpopulations of prostate extracellular vesicles in patient plasma. Prostate 2019, 79, 592–603.
- . Higginbotham, J.N.; Zhang, Q.; Jeppesen, D.K.; Scott, A.M.; Manning, H.C.; Ochieng, J.; Franklin, J.L.; Coffey, R.J. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell. Vesicles 2016, 5, 29254.
- Mastoridis, S.; Bertolino, G.M.; Whitehouse, G.; Dazzi, F.; Sanchez-Fueyo, A.; Martinez-Llordella, M. Multiparametric Analysis of Circulating Exosomes and Other Small Extracellular Vesicles by Advanced Imaging Flow Cytometry. Front. Immunol. 2018, 9, 1583.
- Löf, L.; Arngården, L.; Ebai, T.; Landegren, U.; Söderberg, O.; Kamali-Moghaddam, M. Detection of Extracellular Vesicles Using Proximity Ligation Assay with Flow Cytometry Readout-ExoPLA. Curr. Protoc. Cytom. 2017, 81, 4–8.
- Larssen, P.; Wik, L.; Czarnewski, P.; Eldh, M.; Löf, L.; Ronquist, K.G.; Dubois, L.; Freyhult, E.; Gallant, C.J.; Oelrich, J.; et al. Tracing Cellular Origin of Human Exosomes Using Multiplex Proximity Extension Assays. Mol. Cell Proteom. 2017, 16, 1547, Erratum in Mol. Cell. Proteom. 2017, 16, 502–511.
- Löf, L.; Ebai, T.; Dubois, L.; Wik, L.; Ronquist, K.G.; Nolander, O.; Lundin, E.; Söderberg, O.; Landegren, U.; Kamali-Moghaddam, M. Detecting individual extracellular vesicles using a multicolor in situ proximity ligation assay with flow cytometric readout. Sci. Rep. 2016, 6, 34358.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.8K
Entry Collection:
Extraction Techniques in Sample Preparation
Revisions:
4 times
(View History)
Update Date:
20 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




