
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Tania Ho | -- | 5079 | 2022-07-04 12:24:09 | | | |
| 2 | Jessie Wu | -2495 word(s) | 2584 | 2022-07-05 03:50:36 | | | | |
| 3 | Jessie Wu | Meta information modification | 2584 | 2022-07-05 03:56:01 | | | | |
| 4 | Jessie Wu | -17 word(s) | 2567 | 2022-07-05 03:57:59 | | | | |
| 5 | Jessie Wu | Meta information modification | 2567 | 2022-07-05 04:02:22 | | | | |
| 6 | Jessie Wu | + 2 word(s) | 2569 | 2022-07-05 04:03:01 | | |
Video Upload Options
The cooperative and mutualistic interaction between plant roots and arbuscular mycorrhizal (AM) fungi from the phylum Glomeromycota is tightly regulated by both partners at the cellular, molecular and genetic levels, and it is highly dependent on environmental and biological variables.
1. Nutritional Regulation
The main sense and the success of the arbuscular mycorrhizal (AM) symbiosis basically resides in the mutual nutritional benefits obtained by both interacting partners, the plant and the AM fungus. Plants provide carbon (C) to AM fungi in form of carbohydrates and fatty acids, while AM fungi provide minerals, mainly inorganic phosphate (Pi) and nitrogen to the plants. It is logical to assume that the availability of the different nutrients is a key factor determining the establishment and development of the interaction.
Among the different exchanged nutrients, phosphate is the one that is thought to be the most important in the regulation of the AM symbiosis. Actually, high phosphate supply causes inhibition of root colonization from a very early stage, including hyphopodia formation [1][2], and high Pi negatively affects presymbiotic signalling [3]. Although less studied, nitrogen is also a nutritional determinant of the interaction. Nouri, et al.[4] have shown that N starvation reduces the negative effect of high Pi on the AM symbiosis, and premature arbuscule degeneration observed in plants mutated for genes encoding specific mycorrhizal Pi transporters is prevented at low N conditions in the media. Although researchers cannot discard a regulatory role of other mineral nutrients, they may have a minor importance in comparison to Pi and N. For example, in petunia plants inoculated with Rhizophagus irregularis, no AM development-sensitivity was detected for any of the alternative tested nutrients, including sulfate, calcium, magnesium, and iron [4].
Carbon availability (in form of sugars and lipids) from the plant to the AM fungus is also a factor determining AM development. In soybean plants, it has been shown that inoculation with AM fungus species exhibiting higher colonization rates, compared to those ones with lower AM colonization, is accompanied with an increased plant growth and with higher sugar contents, as well as a higher induction of genes related to sugar metabolism and transport [5]. Recent research has also revealed a regulatory role of lipids in the AM symbiosis. The reduced lipid transfer from hosts to AM fungi inhibits arbuscule formation [6].
2. Arbuscular Mycorrhizal Regulation by Hormones and Other Signalling Molecules
Clear evidence shows that AM symbiosis alters plant hormonal homeostasis and almost all phytohormones have a key regulatory role in the establishment and functionality of the AM symbiosis, as shown by many studies based on the application of hormone treatments or on the analysis of plants with alterations in hormone biosynthesis or signalling. Plant hormones have been reported to act from early stages in the presymbiotic signalling, to later stages (revised by [Pozo, et al. [7], Bedini, et al. [8], Liao, et al. [9]).
Before the physical contact between the AM fungus and the plant root, it is required a molecular communication between both symbionts. The key elements for this communication are strigolactones (SLs) from the plant side. Once these compounds are secreted from the root to the rhizosphere in response to Pi deficiency conditions [10][11], they serve as signals indicating the presence of a host receptive to be colonized by AM fungi. In this sense, many reports show that mycorrhizal colonization is significantly reduced in plant mutants defective for the biosynthesis and export of strigolactones [10][12][13][14]. Abscisic acid (ABA) is another apocarotenoid hormone with a regulatory role in mycorrhizal root colonization. The ABA sitiens tomato mutants with reduced ABA concentrations showed a reduced AM colonization and a lower percentage of well-developed arbuscules [15][16], while mycorrhizal colonization and arbuscule intensity were promoted in ABA pre-treated potato plants [17]. Charpentier, et al.[18] showed a dual role of ABA on AM symbiosis in Medicago. Several reports suggest that the giberellin-DELLA complex also plays an essential role in the control of the symbiosis. Mycorrhizal development is ligated to the increased levels of GAs, and an increased expression of genes associated to their biosynthesis [19][20][21]. Experiments with pea mutants deficient in gibberelins, as well as exogenous GA treatment applied to rice and tomato plants support a negative regulatory role of GAs on mycorrhizal colonization [20][22][23]. Brassinosteroids are also thought to have a signalling role during AM symbiosis. Actually, a reduced mycorrhizal colonization is observed in mutant plants impaired in the brassinoesteroid receptor or in brassinoesteroid biosynthesis [24][25]. Although the role of other plant hormones in mycorrhizal establishment and development has been less studied, it is thought that most of them are involved in these processes. Salicylic acid, ethylene and cytokinins have been reported to have a negative role on AM fungal penetration and colonization [22], while auxins have been observed to positively regulate arbuscule development and functionality [26]. For the jasmonic acid, both positive and negative effects on mycorrhization have been observed [27]. Moreover, as it is well-known, hormones do not act independently, but a complex hormonal dialog regulates plant development and responses. In the case of AM regulation, the antagonistic interactions ABA-ethylene and ABA-giberellin, have been shown to regulate mycorrhizal development and arbuscule formation, respectively [16][28][29].
In addition to the hormonal signals, there is evidence indicating the presence of other essential signalling molecules during mycorrhization. For example, recent insights point to a role of coumarins as novel signals in the pre-symbiotic chemical dialog by promoting fungal metabolism and inducing the initial steps of AM colonization [30]. Another candidate to have a role in pre-symbiotic signalling is a hypothetical compound transported by the plant N-acetylglucosamine exporter NOPE1 [31]. In addition, recent research has shown that the KAI2/D14L and the DLK2 receptors, which are phylogenetically close to the strigolactone receptor D14, play relevant roles in the mycorrhizal symbiosis [32][33], and then it is expected that the not-yet identified corresponding ligands (of plant or fungal origin) might be important in AM signalling and regulation.
Apocarotenoid compounds seem to be especially important in AM regulation [34]. Apart from SLs and ABA, and the possible ligands of D14L and DLK2, other types of apocarotenoid molecules such as zaxinone, cyclohexanone derivatives and mycorrhizadicins are very likely involved in the control of the AM symbiosis [35][36][37][38].
Knowledge about the possible signals from fungal origin involved in the regulation of mycorrhization is very scarce. Due to methodological limitations to perform genetic approaches on AM fungi, it is very difficult to assign specific signalling roles to a particular fungal molecule. At this respect, a combination of two kinds of chitinaceous molecules, commonly known as Myc factors, has been shown to be essential for AM establishment [39]: the Myc-LCOs (lipochitooligosaccharides) and the Myc-COs (short-chain chitin oligomers). AM fungi also secrete proteins, known as “effectors” to communicate with the host plant and to modulate the immune response to allow mycorrhization. Although only SP7, SIS1 and CRN1 have been identified as fungal effectors participating in the establishment of the AM association [40][41][42], it is expected that many other effectors should be involved in AM regulation. Actually, in silico analyses in the genome of R. proliferus have predicted the presence of coding regions for 416 small secreted peptides [43]. Also, in the genome of R. irregularis, 220 candidate effector genes are present, of which 95% are also found in R. clarus.
3. Transcriptional Regulation of Arbuscular Mycorrhizal Symbiosis
A considerable number of studies have shown that large transcriptional changes are induced in the plant host during all stages of colonization. The major portion of regulated genes is involved in signalling, protein metabolism, nutrient transport, secondary metabolite biosynthesis, cell wall modification and lipid metabolism. Furthermore, a significant number of genes encoding putative transcriptional regulators are differentially expressed in mycorrhizal roots, suggesting that AM development is regulated by a complex transcriptional control network [44][45][46][47] in which the GRAS gene family have a prominent role [48] (Figure 1).
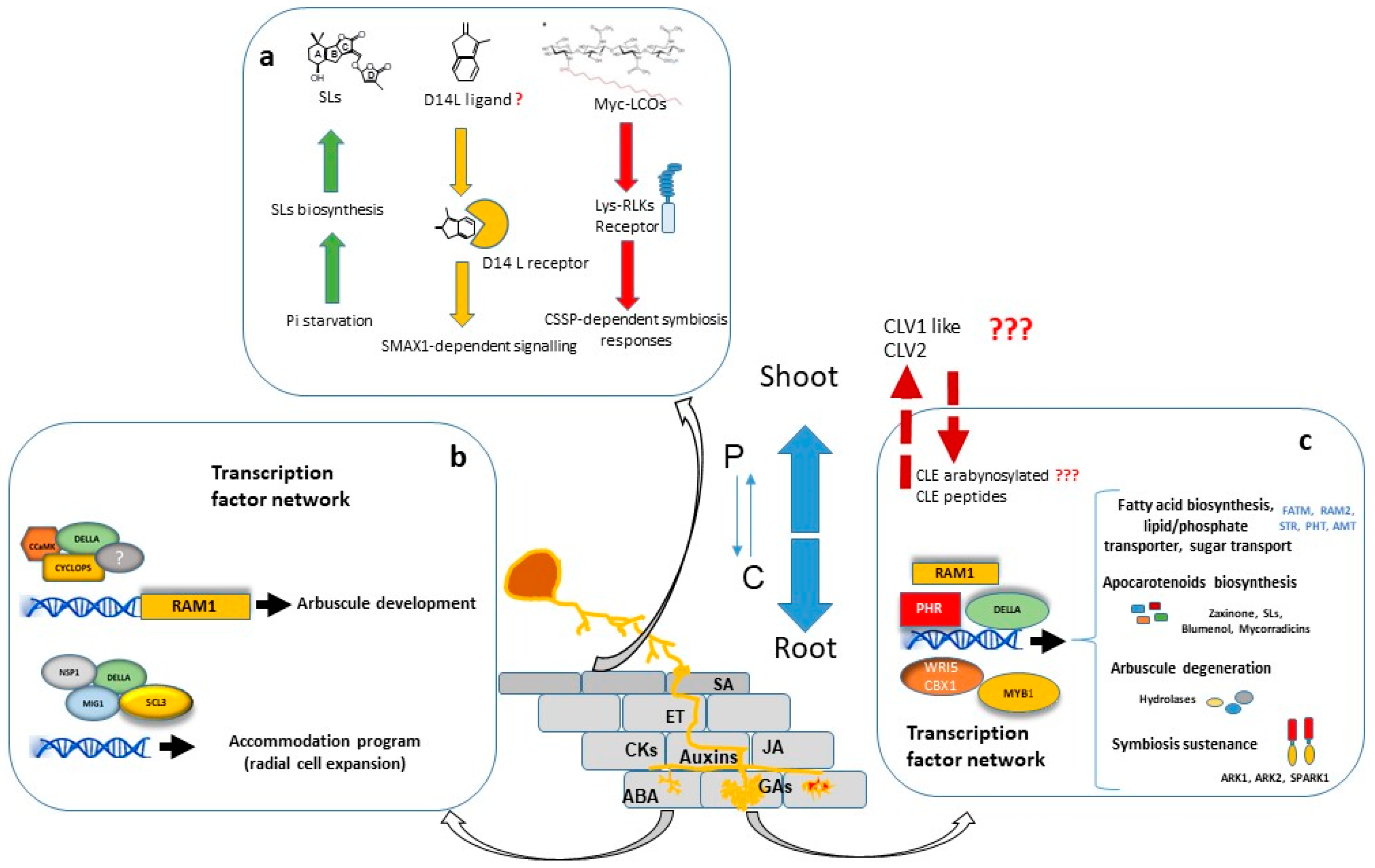
Transcriptional regulation of symbiotic genes is in part dependent of the Common Symbiosis Signalling Pathway (CSSP) activated during AM and Root Nodulation. Nuclear calcium oscillations generated in plant root cells upon the perception of external symbiotic fungal signals, activate CYCLOPS/IPD3, a primary and central transcription factor of the symbiotic signalling response [49][50]. Among other possible direct target promoters, CYCLOPS in a complex with CCaMK and DELLA, binds the RAM1 promoter and induces RAM1 expression [51], which encodes a key transcription factor required for arbuscule development. Part of the Myc-LCO and CO response is dependent on the GRAS protein NSP1 [52][53][54], probably by establishing a regulatory module together with NSP2 and the CYCLOPS-CCaMK-DELLA complex, as suggested by Jin, et al. [55].
Interestingly, recent research has shown that transcriptional regulation of symbiotic genes is not only dependent on the CSSP triggered upon perception of fungal signals and mediated by CYCLOPS/IPD3, but also the phosphate starvation signalling plays a highly relevant role, and the PHR (Phosphate starvation response) TFs govern the regulation of AM-related genes [56]. Moreover, computational analysis carried out by these authors revealed that 42% of the promoter regions of AM-regulated genes in rice carry P1BS (PHR1 Biding Site) motifs, strongly suggesting that Pi starvation plays a central role in the transcriptional activation of a wide range of AM-symbiotic genes.
For arbuscule development, RAM1 is assumed to be the master transcriptional regulator. RAM1 gene expression is induced by both, Pi starvation conditions through binding of PHR TF to the P1BS element of the RAM1 promoter [56], and also by the CSSP through binding of CYCLOPS TF to a cis element (AMCYC-RE) of the RAM1 promoter [51]. Experiments performed in M. truncatula, L. japonicus and petunia, suggest that RAM1 is essential for the formation of the periarbuscular membrane and for arbuscule branching, and also probably for plant-fungus nutrient exchange [51][57][58][59][60]. The RAM1-dependent activation of several of these symbiotic genes might be mediated by AP2 transcription factors, such as WRI5 or CBX1 [59][61][62].
Arbuscule development requires to be accompanied by cell expansion of cortex cells for accommodation of arbuscules. At this regard, the adjustment of cell size during arbuscule life cycle has been suggested to be regulated by two different modules of GRAS transcription factors with antagonistic actions. In one hand, MIG1 (Mycorrhiza Induced GRAS 1) transcription factor, in a complex with DELLA, promotes radial expansion of arbuscule hosting cells while, in the other hand, MIG2 and SCL3, also in concert with DELLA, restrict cell expansion [63][64].
Arbuscules are continuously being recycled. They have a relatively short life, around 2-3 days, and are rapidly degraded after 2-7 days [65][66]. The quick removal of senescent arbuscules might be a plant regulatory mechanism to restrict the presence of fungal arbuscules that are not providing benefits to the plant. The M. truncatula AM-induced gene MYB1 encodes a transcription factor which, in association with NSP1 and DELLA, has been reported as a key regulatory element required for the induction of many genes associated to arbuscule degeneration, such as cysteine proteases and chitinases [67]. Recent research carried out by Wang, et al. [68] in M. truncatula, also shows that the SPX-domain containing proteins SPX1 and SPX3 regulate arbuscule degradation, probably by sensing the delivered Pi at the arbuscule level.
Apart from the transcriptional regulation mediated by TFs, posttranscriptional mechanisms for AM gene expression regulation have been also identified. For example, miRNAs from the miR171 family seem to be involved in maintaining the balance of AM colonization [69][70]. In particular, the microRNA miR171h, which is induced in M. truncatula during AM colonization and by Myc-LCOs, has a repressing role on mycorrhizal colonization [69].
4. Systemic Autoregulation of Arbuscular Mycorrhizal Symbiosis
In order to balance the energy cost with the benefit gained, plants employ a systemic negative feedback loop to control the formation of nutrient-acquiring symbioses. This mechanism of feedback control is particularly important in legumes, where existing nodules systemically inhibit subsequent nodulation in other parts of the root system through a process termed “autoregulation of nodulation” (AON) [71]. A similar regulatory mechanism for AM symbiosis has been reported, and the systemic autoregulation of AM colonization (AOM) in split root studies has been observed in both legumes and non-legumes [72][73]. AOM pathway shares some elements with AON, and physiological studies in legumes have indicated there is at least some overlap in the genes and signals that regulate these two symbioses. In addition to its role in nodulation, the CLV1-like protein is also essential in the AOM pathway since clv1-like mutants across legume species (sym29, sunn, nark, and har1) also display an increased AM fungal colonization, implicating a role for these LRR-RLKs in autoregulation of mycorrhizal symbiosis (revised by Wang, et al. [74]). Recently, first genetic evidence for the AOM pathway in non-legumes has been obtained in tomato. Compared with WT, clv2 plants displayed a significant increase in AM colonization, including arbuscule frequency, suggesting a role for the tomato CLV2 in AM development [74]. In addition, recent studies provided evidence for a functional role of CLE-mediated signalling in AOM [75][76].
References
- Balzergue, C.; Puech-Pagès, V.; Bécard, G.; Rochange, S. F., The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. J. Exp. Bot. 2011, 62, (3), 1049-1060.
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; Kuhlemeier, C.; Martinoia, E.; Franken, P.; Scholz, U.; Reinhardt, D., Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010, 64, (6), 1002-17.
- Russo, G.; Spinella, S.; Sciacca, E.; Bonfante, P.; Genre, A., Automated analysis of calcium spiking profiles with CaSA software: two case studies from root-microbe symbioses. BMC Plant Biol. 2013, 13, (1), 224.
- Nouri, E.; Breuillin-Sessoms, F.; Feller, U.; Reinhardt, D., Phosphorus and nitrogen regulate arbuscular mycorrhizal symbiosis in Petunia hybrida. PloS one 2014, 9, (3), e90841.
- Zhao, S.; Chen, A.; Chen, C.; Li, C.; Xia, R.; Wang, X., Transcriptomic analysis reveals the possible roles of sugar metabolism and export for positive mycorrhizal growth responses in soybean. Physiol. Plant. 2019, 166, (3), 712-728.
- Feng, Z.; Liu, X.; Feng, G.; Zhu, H.; Yao, Q., Linking lipid transfer with reduced arbuscule formation in tomato roots colonized by arbuscular mycorrhizal fungus under low pH stress. Environ. Microbiol. 2020, 22, (3), 1036-1051.
- Pozo, M. J.; López‐Ráez, J. A.; Azcón‐Aguilar, C.; García‐Garrido, J. M., Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015, 205, (4), 1431-1436.
- Bedini, A.; Mercy, L.; Schneider, C.; Franken, P.; Lucic-Mercy, E., Unraveling the initial plant hormone signaling, metabolic mechanisms and plant defense triggering the endomycorrhizal symbiosis behavior. Front. Plant Sci. 2018, 9, 1800.
- Liao, D.; Wang, S.; Cui, M.; Liu, J.; Chen, A.; Xu, G., Phytohormones regulate the development of arbuscular mycorrhizal symbiosis. International journal of molecular sciences 2018, 19, (10), 3146.
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J. B.; Reinhardt, D.; Bours, R.; Bouwmeester, H. J.; Martinoia, E., A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, (7389), 341.
- Yoneyama, K.; Yoneyama, K.; Takeuchi, Y.; Sekimoto, H., Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 2007, 225, (4), 1031-1038.
- Gomez-Roldan, V.; Fermas, S.; Brewer, P. B.; Puech-Pagès, V.; Dun, E. A.; Pillot, J.-P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.-C., Strigolactone inhibition of shoot branching. Nature 2008, 455, (7210), 189.
- Koltai, H.; LekKala, S. P.; Bhattacharya, C.; Mayzlish-Gati, E.; Resnick, N.; Wininger, S.; Dor, E.; Yoneyama, K.; Yoneyama, K.; Hershenhorn, J., A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 2010, 61, (6), 1739-1749.
- Yoshida, S.; Kameoka, H.; Tempo, M.; Akiyama, K.; Umehara, M.; Yamaguchi, S.; Hayashi, H.; Kyozuka, J.; Shirasu, K., The D3 F‐box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 2012, 196, (4), 1208-1216.
- Herrera‐Medina, M. J.; Steinkellner, S.; Vierheilig, H.; Ocampo Bote, J. A.; García Garrido, J. M., Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 2007, 175, (3), 554-564.
- Martín‐Rodríguez, J. Á.; León‐Morcillo, R.; Vierheilig, H.; Ocampo, J. A.; Ludwig‐Müller, J.; García‐Garrido, J. M., Ethylene‐dependent/ethylene‐independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol. 2011, 190, (1), 193-205.
- Mercy, L.; Lucic-Mercy, E.; Nogales, A.; Poghosyan, A.; Schneider, C.; Arnholdt-Schmitt, B., A functional approach towards understanding the role of the mitochondrial respiratory chain in an endomycorrhizal symbiosis. Front. Plant Sci. 2017, 8, 417.
- Charpentier, M.; Sun, J.; Wen, J.; Mysore, K. S.; Oldroyd, G. E., Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol. 2014, 166, (4), 2077-2090.
- García Garrido, J. M.; León Morcillo, R. J.; Martín Rodríguez, J. A.; Ocampo Bote, J. A., Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol. Plant-Microbe Interact. 2010, 23, (5), 651-64.
- Martín‐Rodríguez, J. Á.; Ocampo, J. A.; Molinero‐Rosales, N.; Tarkowská, D.; Ruíz‐Rivero, O.; García‐Garrido, J. M., Role of gibberellins during arbuscular mycorrhizal formation in tomato: new insights revealed by endogenous quantification and genetic analysis of their metabolism in mycorrhizal roots. Physiol. Plant. 2015, 154, (1), 66-81.
- Takeda, N.; Handa, Y.; Tsuzuki, S.; Kojima, M.; Sakakibara, H.; Kawaguchi, M., Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus. Plant Physiol. 2015, 167, (2), 545-557.
- Foo, E.; Ross, J. J.; Jones, W. T.; Reid, J. B., Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann. Bot. 2013, 111, (5), 769-779.
- Yu, N.; Luo, D.; Zhang, X.; Liu, J.; Wang, W.; Jin, Y.; Dong, W.; Liu, J.; Liu, H.; Yang, W., A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2014, 24, (1), 130.
- Bitterlich, M.; Krügel, U.; Boldt-Burisch, K.; Franken, P.; Kühn, C., Interaction of brassinosteroid functions and sucrose transporter SlSUT2 regulate the formation of arbuscular mycorrhiza. Plant Signal Behav 2014, 9, (10), e970426.
- Bitterlich, M.; Krügel, U.; Boldt‐Burisch, K.; Franken, P.; Kühn, C., The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 2014, 78, (5), 877-889.
- Etemadi, M.; Gutjahr, C.; Couzigou, J.-M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Bécard, G.; Combier, J.-P., Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014, 166, (1), 281-292.
- Wasternack, C.; Hause, B., Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, (6), 1021-1058.
- Gutjahr, C., Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 2014, 20, 26-34.
- Martin-Rodriguez, J. A.; Huertas, R.; Ho-Plagaro, T.; Ocampo, J. A.; Tureckova, V.; Tarkowska, D.; Ludwig-Muller, J.; Garcia-Garrido, J. M., Gibberellin-Abscisic Acid Balances during Arbuscular Mycorrhiza Formation in Tomato. Frontiers in plant science 2016, 7, 1273.
- Cosme, M.; Fernández, I.; Declerck, S.; van der Heijden, M. G.; Pieterse, C. M., A coumarin exudation pathway mitigates arbuscular mycorrhizal incompatibility in Arabidopsis thaliana. Plant Mol. Biol. 2021, 1-16.
- Nadal, M.; Sawers, R.; Naseem, S.; Bassin, B.; Kulicke, C.; Sharman, A.; An, G.; An, K.; Ahern, K. R.; Romag, A., An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 2017, 3, (6), 17073.
- Gutjahr, C.; Gobbato, E.; Choi, J.; Riemann, M.; Johnston, M. G.; Summers, W.; Carbonnel, S.; Mansfield, C.; Yang, S. Y.; Nadal, M.; Acosta, I.; Takano, M.; Jiao, W. B.; Schneeberger, K.; Kelly, K. A.; Paszkowski, U., Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 2015, 350, (6267), 1521-4.
- Ho‐Plágaro, T.; Morcillo, R. J.; Tamayo‐Navarrete, M. I.; Huertas, R.; Molinero‐Rosales, N.; López‐Ráez, J. A.; Macho, A. P.; García‐Garrido, J. M., DLK2 regulates arbuscule hyphal branching during arbuscular mycorrhizal symbiosis. New Phytol. 2021, 229, (1), 548-562.
- Fiorilli, V.; Wang, J. Y.; Bonfante, P.; Lanfranco, L.; Al-Babili, S., Apocarotenoids: old and new mediators of the arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2019, 1186.
- Wang, J. Y.; Haider, I.; Jamil, M.; Fiorilli, V.; Saito, Y.; Mi, J.; Baz, L.; Kountche, B. A.; Jia, K.-P.; Guo, X., The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 2019, 10, (1), 1-9.
- Fester, T.; Hause, B.; Schmidt, D.; Halfmann, K.; Schmidt, J.; Wray, V.; Hause, G.; Strack, D., Occurrence and localization of apocarotenoids in arbuscular mycorrhizal plant roots. Plant Cell Physiol. 2002, 43, (3), 256-265.
- Fester, T.; Maier, W.; Strack, D., Accumulation of secondary compounds in barley and wheat roots in response to inoculation with an arbuscular mycorrhizal fungus and co-inoculation with rhizosphere bacteria. Mycorrhiza 1999, 8, (5), 241-246.
- Floss, D. S.; Schliemann, W.; Schmidt, J.; Strack, D.; Walter, M. H., RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol. 2008, 148, (3), 1267-1282.
- Feng, F.; Sun, J.; Radhakrishnan, G. V.; Lee, T.; Bozsóki, Z.; Fort, S.; Gavrin, A.; Gysel, K.; Thygesen, M. B.; Andersen, K. R., A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat. Commun. 2019, 10, (1), 1-12.
- Kloppholz, S.; Kuhn, H.; Requena, N., A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 2011, 21, (14), 1204-1209.
- Tsuzuki, S.; Handa, Y.; Takeda, N.; Kawaguchi, M., Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol. Plant-Microbe Interact. 2016, 29, (4), 277-286.
- Voß, S.; Betz, R.; Heidt, S.; Corradi, N.; Requena, N., RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Front. Microbiol. 2018, 9, 2068.
- Prasad Singh, P.; Srivastava, D.; Jaiswar, A.; Adholeya, A., Effector proteins of Rhizophagus proliferus: conserved protein domains may play a role in host-specific interaction with different plant species. Braz. J. Microbiol. 2019, 50, (3), 593-601.
- Rich, M. K.; Courty, P. E.; Roux, C.; Reinhardt, D., Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genomics 2017, 18, (1), 589.
- Xue, L.; Cui, H.; Buer, B.; Vijayakumar, V.; Delaux, P.-M.; Junkermann, S.; Bucher, M., Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 2015, 167, (3), 854-871.
- Hartmann, R. M.; Schaepe, S.; Nübel, D.; Petersen, A. C.; Bertolini, M.; Vasilev, J.; Küster, H.; Hohnjec, N., Insights into the complex role of GRAS transcription factors in the arbuscular mycorrhiza symbiosis. Scientific reports 2019, 9, (1), 1-15.
- Ho-Plágaro, T.; Molinero-Rosales, N.; Flores, D. F.; Díaz, M. V.; García-Garrido, J. M., Identification and expression analysis of GRAS transcription factor genes involved in the control of arbuscular mycorrhizal development in tomato. Front. Plant Sci. 2019, 10, 268.
- García-Garrido, J. M.; Ho Plágaro, T., Multifarious and interactive roles of GRAS transcription factors during arbuscular mycorrhiza development. Front. Plant Sci. 2022, 13, 908.
- Yano, K.; Yoshida, S.; Müller, J.; Singh, S.; Banba, M.; Vickers, K.; Markmann, K.; White, C.; Schuller, B.; Sato, S., CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. 2008, 105, (51), 20540-20545.
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M., CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell host & microbe 2014, 15, (2), 139-152.
- Pimprikar, P.; Carbonnel, S.; Paries, M.; Katzer, K.; Klingl, V.; Bohmer, M. J.; Karl, L.; Floss, D. S.; Harrison, M. J.; Parniske, M., A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 2016, 26, (8), 987-998.
- Camps, C.; Jardinaud, M. F.; Rengel, D.; Carrère, S.; Hervé, C.; Debellé, F.; Gamas, P.; Bensmihen, S.; Gough, C., Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc‐LCOs in Medicago truncatula. New Phytol. 2015, 208, (1), 224-240.
- Sun, J.; Miller, J. B.; Granqvist, E.; Wiley-Kalil, A.; Gobbato, E.; Maillet, F.; Cottaz, S.; Samain, E.; Venkateshwaran, M.; Fort, S., Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 2015, 27, (3), 823-838.
- Delaux, P. M.; Bécard, G.; Combier, J. P., NSP1 is a component of the Myc signaling pathway. New Phytol. 2013, 199, (1), 59-65.
- Jin, Y.; Liu, H.; Luo, D.; Yu, N.; Dong, W.; Wang, C.; Zhang, X.; Dai, H.; Yang, J.; Wang, E., DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016, 7.
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Xie, Q.; Wang, G.; Xiao, Y.; Chen, F., A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, (22), 5527-5540. e18.
- Gobbato, E.; Marsh, J. F.; Vernié, T.; Wang, E.; Maillet, F.; Kim, J.; Miller, J. B.; Sun, J.; Bano, S. A.; Ratet, P., A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 2012, 22, (23), 2236-2241.
- Park, H.-J.; Floss, D. S.; Levesque-Tremblay, V.; Bravo, A.; Harrison, M. J., Hyphal branching during arbuscule development requires RAM1. Plant Physiol. 2015, pp. 01155.2015.
- Luginbuehl, L. H.; Menard, G. N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G. V.; Breakspear, A.; Oldroyd, G. E.; Eastmond, P. J., Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, eaan0081.
- Rich, M. K.; Schorderet, M.; Bapaume, L.; Falquet, L.; Morel, P.; Vandenbussche, M.; Reinhardt, D., The petunia GRAS transcription factor ATA/RAM1 regulates symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol. 2015, 168, (3), 788-797.
- Jiang, Y.; Xie, Q.; Wang, W.; Yang, J.; Zhang, X.; Yu, N.; Zhou, Y.; Wang, E., Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol. plant 2018, 11, (11), 1344-1359.
- Xue, L.; Klinnawee, L.; Zhou, Y.; Saridis, G.; Vijayakumar, V.; Brands, M.; Dörmann, P.; Gigolashvili, T.; Turck, F.; Bucher, M., AP2 transcription factor CBX1 with a specific function in symbiotic exchange of nutrients in mycorrhizal Lotus japonicus. Proc. Natl. Acad. Sci. 2018, 115, (39), E9239-E9246.
- Heck, C.; Kuhn, H.; Heidt, S.; Walter, S.; Rieger, N.; Requena, N., Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 2016, 26, (20), 2770-2778.
- Seemann, C.; Heck, C.; Voß, S.; Schmoll, J.; Enderle, E.; Schwarz, D.; Requena, N., Root cortex development is fine‐tuned by the interplay of MIGs, SCL3 and DELLAs during arbuscular mycorrhizal symbiosis. New Phytol. 2022, 233, (2), 948-965.
- Kobae, Y.; Hata, S., Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 2010, 51, (3), 341-53.
- Alexander, T.; Toth, R.; Meier, R.; Weber, H. C., Dynamics of arbuscule development and degeneration in onion, bean, and tomato with reference to vesicular–arbuscular mycorrhizae in grasses. Can. J. Bot. 1989, 67, (8), 2505-2513.
- Floss, D. S.; Gomez, S. K.; Park, H.-J.; MacLean, A. M.; Müller, L. M.; Bhattarai, K. K.; Lévesque-Tremblay, V.; Maldonado-Mendoza, I. E.; Harrison, M. J., A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr. Biol. 2017, 27, (8), 1206-1212.
- Wang, P.; Snijders, R.; Kohlen, W.; Liu, J.; Bisseling, T.; Limpens, E., Medicago SPX1 and SPX3 regulate phosphate homeostasis, mycorrhizal colonization, and arbuscule degradation. Plant Cell 2021, 33, (11), 3470-3486.
- Lauressergues, D.; Delaux, P. M.; Formey, D.; Lelandais‐Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J. P., The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012, 72, (3), 512-522.
- Couzigou, J.-M.; Lauressergues, D.; André, O.; Gutjahr, C.; Guillotin, B.; Bécard, G.; Combier, J.-P., Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell host & microbe 2017, 21, (1), 106-112.
- Caetano-Anollés, G.; Gresshoff, P. M., Plant genetic control of nodulation. Annu. Rev. Microbiol. 1991, 45, (1), 345-382.
- Vierheilig, H.; Maier, W.; Wyss, U.; Samson, J.; Strack, D.; Piché, Y., Cyclohexenone derivative-and phosphate-levels in split-root systems and their role in the systemic suppression of mycorrhization in precolonized barley plants. J. Plant Physiol. 2000, 157, (6), 593-599.
- Meixner, C.; Vegvari, G.; Ludwig‐Müller, J.; Gagnon, H.; Steinkellner, S.; Staehelin, C.; Gresshoff, P.; Vierheilig, H., Two defined alleles of the LRR receptor kinase GmNARK in supernodulating soybean govern differing autoregulation of mycorrhization. Physiol. Plant. 2007, 130, (2), 261-270.
- Wang, C.; Reid, J. B.; Foo, E., The art of self-control–autoregulation of plant–microbe symbioses. Front. Plant Sci. 2018, 9, 988.
- Müller, L. M.; Flokova, K.; Schnabel, E.; Sun, X.; Fei, Z.; Frugoli, J.; Bouwmeester, H. J.; Harrison, M. J., A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants 2019, 5, (9), 933-939.
- Karlo, M.; Boschiero, C.; Landerslev, K. G.; Blanco, G. S.; Wen, J.; Mysore, K. S.; Dai, X.; Zhao, P. X.; de Bang, T. C., The CLE53–SUNN genetic pathway negatively regulates arbuscular mycorrhiza root colonization in Medicago truncatula. J. Exp. Bot. 2020, 71, (16), 4972-4984.




