Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Maria Luz Martinez-Chantar | -- | 2022 | 2022-06-22 15:35:18 | | | |
| 2 | Nora Tang | Meta information modification | 2022 | 2022-06-24 03:31:35 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lachiondo-Ortega, S.; Delgado, T.C.; Baños-Jaime, B.; Velázquez-Cruz, A.; Díaz-Moreno, I.; Martínez-Chantar, M.L. HuR and Hepatobiliary Cancers. Encyclopedia. Available online: https://encyclopedia.pub/entry/24355 (accessed on 08 February 2026).
Lachiondo-Ortega S, Delgado TC, Baños-Jaime B, Velázquez-Cruz A, Díaz-Moreno I, Martínez-Chantar ML. HuR and Hepatobiliary Cancers. Encyclopedia. Available at: https://encyclopedia.pub/entry/24355. Accessed February 08, 2026.
Lachiondo-Ortega, Sofia, Teresa Cardoso Delgado, Blanca Baños-Jaime, Alejandro Velázquez-Cruz, Irene Díaz-Moreno, María Luz Martínez-Chantar. "HuR and Hepatobiliary Cancers" Encyclopedia, https://encyclopedia.pub/entry/24355 (accessed February 08, 2026).
Lachiondo-Ortega, S., Delgado, T.C., Baños-Jaime, B., Velázquez-Cruz, A., Díaz-Moreno, I., & Martínez-Chantar, M.L. (2022, June 22). HuR and Hepatobiliary Cancers. In Encyclopedia. https://encyclopedia.pub/entry/24355
Lachiondo-Ortega, Sofia, et al. "HuR and Hepatobiliary Cancers." Encyclopedia. Web. 22 June, 2022.
Copy Citation
HuR owns the post-transcriptional control of a large number of RNAs, enabling the protein to play pivotal roles that are dictated by the molecular functions of the transcripts. HuR targets, chiefly, include many mRNAs encoding proteins involved in cell proliferation, senescence, apoptosis, differentiation, stress, and immune responses. In turn, HuR was found to be involved not only in physiological processes (e.g., adipogenesis and muscle differentiation) but also in disease (primarily, cancer and inflammation).
ELAV-like protein 1
RNA-binding protein
hepatocellular carcinoma
cholangiocarcinoma
1. Hepatobiliary Cancers
Hepatobiliary cancers are a group of primary malignancies encompassing the liver, the intra- and extra-hepatic biliary tracts, and the gall bladder. Within the liver, the most common primary cancer is hepatocellular carcinoma (HCC). HCC, usually, arises in patients with an underlying chronic liver disease, such as chronic infection with the hepatitis B (HBV) or C virus (HBC), cirrhosis, excessive alcohol consumption, metabolic syndrome, diabetes, and non-alcoholic fatty liver disease (NAFLD) [1]. Even though the clinical management of HCC has improved in the past ten years, particularly for patients at advanced stages, HCC is still the third-most common cause of cancer-associated death and the sixth-most prevalent type of cancer worldwide [2]. Of relevance, in the past few years, important studies on immunotherapy in HCC have been carried out, resulting in novel guidelines for its treatment [3][4][5].
On the other hand, cholangiocarcinoma (CCA) encompasses a group of malignancies, arising at any point in the hepatic biliary tree. Biliary tract cancers, including intrahepatic (ICC), perihilar (PCC), and distal cholangiocarcinoma (DCC), as well as gallbladder cancer, are low-incidence malignancies in most high-income countries, but represent a major health problem in endemic areas. Indeed, ICC is the second-leading cause of primary liver cancer [6]. Nowadays, CCA is a fatal cancer, with a survival rate beyond a year of diagnosis inferior to 5% [7]. Therapeutic options are limited and surgery is the cornerstone of cure for CCA, even though most patients present with locally advanced or metastatic disease [8]. In the past few years, druggable alterations such as fibroblast growth factor receptor 2 (FGFR2) gene fusions and rearrangements, or isocitrate dehydrogenase-1 (IDH-1) and BRAF mutations, have been widely described in CCA patients, further indicating the important differences between iCCA and PCC/DCC, as thoroughly reviewed in [9].
2. HuR as a Prognostic and Diagnostic Biomarker in Hepatobiliary Cancers
HuR protein is, either, overexpressed in most human cancers or overactivated, as denoted by its increased cytoplasmic localization and the translation of various mRNAs involved in carcinogenesis. Regarding liver cancer, Embade et al. [10], reported significantly higher HuR protein levels in the mouse liver progenitor 29 (MLP29) cell line and in the S-adenosylmethionine-deficient (SAMe-D) cell line, isolated from the methionine adenosyltransferase (MAT)1A knockout (MAT1A-KO) mouse model of HCC, compared with primary mouse hepatocytes. Accordingly, higher HuR expression levels were found in human hepatoma cells (e.g., HepG2, Hep3B, SNU398, SNU449, SNU182, and SNU475), than in normal CRL4020 cells [11]. Moreover, in a human liver cancer tissue microarray (TMA) of 59 liver tissue cores from 44 patients, also by Zhu et al. [11], HCC tumor tissues showed significantly higher overall and cytoplasmic HuR staining, compared to normal liver tissues, and this high HuR staining score correlated with worse survival of patients with early-stage HCC. Furthermore, immunofluorescence analyses of normal versus malignant liver tissue revealed that HuR protein is down-regulated in normal human liver samples and up-regulated in HCC samples of different aetiologies (cirrhotic patients with HCV, alcoholic steatohepatitis, and non-alcoholic steatohepatitis (NASH)), where HuR concentration increased, proportionately, to their transformation status [12]. Finally, the expression of ELAVL1 gene is highly induced in the tumor tissue of a cohort of CCA patients, according to The Cancer Genome Atlas (TCGA) mRNA expression repository [13]. Likewise, high cytoplasmic HuR levels are associated with poor survival in patients with surgically resected CCA, treated with adjuvant gemcitabine-based chemotherapy [14].
In sum, HuR overexpression, along with its cytoplasmic localization, are hallmarks of both HCC and CCA, correlating with disease progression and overall survival. Even though the role of HuR in HCC has been widely investigated, with some studies revealing HuR targets and regulators under these conditions, the function of HuR in CCA remains rather unexplored to date.
3. Signaling Pathways Implicated in HCC Involving HuR
The oncogenic-gene-expression programs that allow cancer cells to develop, survive, proliferate, and colonize other tissues are strongly dependent on post-transcriptional mechanisms [15]. As has been previously described, HuR is highly involved in many types of cancer, including HCC and CCA, and so are the numerous HuR-regulated RNAs, which are known to contribute to the main cancer hallmark functions (i.e., enhanced cell proliferation and survival, elevated local angiogenesis, evasion of immune recognition, facilitated cancer cell invasion, and metastasis). Furthermore, a series of transcriptional, post-transcriptional, and post-translational regulators of HuR function have been found to be altered during HCC. Herein, here aimed to analyze the main regulatory axes involving HuR that are associated with hepatobiliary tumors, which can be classified into those related with cell proliferation, invasion, and metastasis; apoptosis; and autophagy (Figure 1). In the process, some of the previously reported HuR RNA targets and regulators of HuR function have been confirmed in the specific context of HCC, while others have been newly identified in this type of tumor.
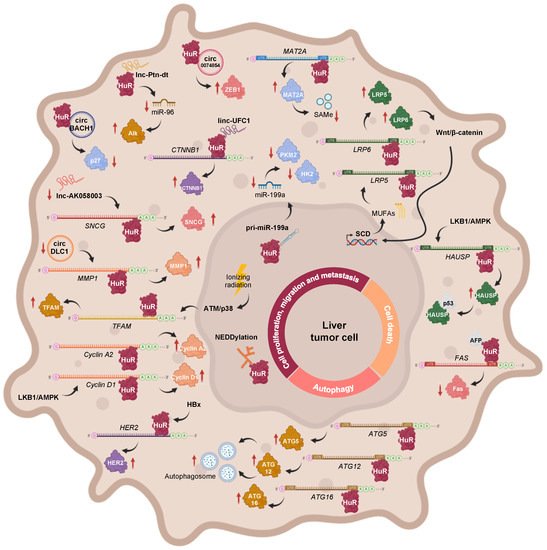
Figure 1. Main regulatory axes involving HuR, which are associated with cell proliferation, invasion, metastasis, apoptosis, and autophagy during HCC. These include the transcriptional, post-transcriptional, and post-translational modulators of HuR function, in addition to HuR target transcripts.
3.1. The Role of HuR in Cell Proliferation, Migration, and Metastasis during HCC
3.1.1. Pathways Involving Coding Transcripts
The PI3K/AKT signaling route is known to be upregulated in HCC [16], and, therefore, it could be responsible for the increased expression of the long HuR mRNA isoform, through NF-kB, as it occurs in gastric tumors [17]. However, it is likely that expression of the short HuR transcript is not observed during HCC, as reduced levels of BMP-7 and p-Smad1/5/8 were detected in patient samples [18].
During HBV-associated HCC, the HBV-encoded X (HBx) protein upregulates HuR expression, which enhances HER2 mRNA stabilization and translation, thus contributing to the migration of HCC cells [19]. In the MAT1A knockout (MAT1A-KO) mouse model, which shows a chronic deficiency in SAMe levels and, spontaneously, develops NASH and HCC, hepatic levels of LKB1 and AMPK are activated, incrementing the cytoplasmic localization of HuR, which leads to the stabilization and expression of cyclin A2 and D1 mRNAs, and subsequent cell cycle progression [20]. Moreover, an abnormally low ratio between methylated and unmethylated HuR was revealed in HCC samples. The two HuR isoforms can associate with the 3′-UTR of MAT2A mRNA, whose activity is linked to liver cell proliferation. However, while unmethylated HuR was shown to stabilize the MAT2A transcript, an increase in its methylation status correlated with lower MAT2A mRNA levels. Hence, the loss of HuR methylation may explain the increased MAT2A mRNA and protein expression, and the subsequent loss of SAMe homeostasis that occurs during hepatocyte dedifferentiation, proliferation, and carcinogenesis [12]. Moreover, related with PTMs, NEDDylation of HuR was firstly reported in the context of liver cancer. Specifically, the E3 ligase Mdm2 catalyzes the conjugation of NEDD8 to HuR at Lys283, Lys313, and Lys326, a process that has been linked to the nuclear localization and reduced proteasomal degradation of the RBP [10]. Interestingly, the Mdm2 transcript being a described target of HuR [21][22], and considering the significantly positive correlation between Mdm2 and HuR expression in clinical HCC and human hepatoma cell lines [10], it would be highly expected that the mRNA levels of this E3 ligase were stabilized by HuR during HCC too, despite not having been verified to date.
The Wnt/β-catenin pathway has been shown to induce stearoyl-CoA desaturase (SCD) expression in liver-tumor-initiating and HCC cells, further increasing the synthesis of mono unsaturated fatty acids (MUFAs). MUFAs can block the nuclear import of HuR, thereby increasing its protein levels in the cytoplasm, where it binds to the 3′UTR of Lrp5 and Lrp6 mRNAs as well as stabilizes and stimulates their translation, further providing a positive feedback loop, by amplifying Wnt/β-catenin signaling and contributing to liver carcinogenesis [23]. Moreover, Wilms tumor 1-associated protein (WTAP) drives N6-methyladenosine (m6A) RNA methylation and epigenetic silencing of ETS1, by interfering with HuR-mediated stabilization of ETS1 mRNA, further alleviating the expression of p21 and p27 G2/M checkpoint proteins, which are known downstream effectors of ETS1, and facilitating HCC progression [24]. Moreover, it has been reported that ionizing radiation activates the DNA damage response (DDR) via ATM/p38, which causes HuR shuttling to the cytoplasm in order to stabilize mitochondrial transcription factor A (TFAM) mRNA and induce its expression in HepG2 hepatoma cells. These results suggest a new pathway, which could be targeted to increase the sensitivity of liver cancer cells to radiotherapy [25].
3.1.2. Pathways Involving Non-Coding Transcripts
In addition to mRNAs, a few examples, whereby HuR interacts with non-coding transcripts during hepatobiliary tumors, have been reported. In human HCC cells subjected to hypoxic stress, HuR binds to the primary transcript of miR-199a (pri-miR-199a) blocking its processing into mature miR-199a. Interestingly, miR-199a is a negative regulator of Hk2 and Pkm2 mRNA expression. Therefore, HuR-mediated miR-199a maturation inhibition, during hypoxia, enables the metabolic reprogramming of HCC cells towards the Warburg effect, which confers favorable conditions for tumor growth, invasion, and metastasis [26]. The long intergenic noncoding RNA (lincRNA)-UFC1 plays an oncogenic role in liver cancer, by interacting with HuR, which stabilizes and induces the expression of the CTNNB1 mRNA, leading to increased cell-cycle progression as well as proliferation and reduced apoptosis in HCC cells [27]. The oncofetal lncRNA Ptn-dt appeared to be highly expressed in HCC tissue and was found to interact with HuR protein, further compromising the stabilization and expression of miR-96. Therefore, the reduced function of miR-96 on the post-transcriptional inhibition of anaplastic lymphoma kinase (Alk) protein contributed to HCC cell proliferation [28]. Another study, describing the tumor suppressor role of lncRNA-AK058003 in HCC, revealed SNCG mRNA as a potential target of HuR. It was postulated that lncRNA-AK058003 is downregulated during HCC but, if overexpressed, it can interact with HuR to suppresses its expression, further affecting SNCG translation and stability, thus inhibiting γ-synuclein-mediated HCC cell proliferation and metastasis, both in vitro and in vivo [29].
In an attempt to elucidate the function of circRNAs, a few circRNA-RBP-mRNA axes involving HuR were revealed in HCC. For example, circBACH1 acts as an oncogene during hepatic tumorigenesis, upon association with HuR, to facilitate its translocation to the cytoplasm, where the RBP inhibits p27 protein expression and allows cell cycle progression, eventually favoring HCC cell proliferation [30]. KIAA1429, a key component of the m6A methyltransferase complex, negatively regulates circRNA-DLC1 in HCC tissues. A mechanistic study revealed that circDLC1 competitively binds with HuR, thereby impairing HuR-mediated MMP1 mRNA stabilization and expression, ultimately resulting in decreased hepatoma cell proliferation and metastasis [31]. Moreover, hsa_circ_0074854 physically interacts and stabilizes HuR protein in the cytoplasm, which induces ZEB1 protein expression, thereby promoting the migration, invasion, and epithelial-mesenchymal transition (EMT) of HepG2 hepatoma cells [32].
3.2. The Role of HuR in Cell Death during HCC
In SAMe-D cells derived from MAT1A KO mice, sustained LKB1 phosphorylation contributes to increased cytoplasmic HuR localization, where it, specifically, binds to the 3′-UTR of herpesvirus-associated ubiquitin-specific protease (HAUSP) mRNA, stabilizing it and increasing its transcription. The subsequent accumulation of HAUSP deubiquitinating enzyme in the cytoplasm allows its interaction with p53, which increases the stability of the tumor suppressor in the cytoplasm, thereby controlling the apoptotic response [33].
A recent study about a possible model, whereby alpha fetoprotein (AFP) regulates HCC progression and chemosensitivity, reported the reactivation of AFP during hepatocarcinogenesis and its interaction with HuR, resulting in the redistribution of the RBP to the cytoplasm. There, HuR would bind to the 3′-UTR of the Fas death receptor mRNA and repress its translation, without affecting its stability or splicing, which further suppresses the Fas/FADD-mediated extrinsic apoptotic program and bypasses immune surveillance in HCC-derived cell lines [11][34].
3.3. The Role of HuR in Autophagy during HCC
Interestingly, HuR binds to the 3′-UTR of ATG5, ATG12, and ATG16 mRNAs and enhances their translation. As a result, autophagosome formation is enhanced, dysregulating the autophagy activity in HCC cell lines, which might possibly act as a pro-survival response and promote hepatic tumor growth [35]. On the other hand, inhibition of autophagy by BECN1 siRNA leads to HuR-enhanced ferroptosis in HCC. HuR and BECN1 interaction induces autophagosome formation, increasing autophagic ferritin degradation and enhancing ferroptosis in hepatic stellate cells (HSCs) [36].
References
- Gerbes, A.; Zoulim, F.; Tilg, H.; Dufour, J.-F.; Bruix, J.; Paradis, V.; Salem, R.; Peck-Radosavljevic, M.; Galle, P.R.; Greten, T.F.; et al. Gut Roundtable Meeting Paper: Selected Recent Advances in Hepatocellular Carcinoma. Gut 2018, 67, 380–388.
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462.
- Rizzo, A.; Ricci, A.D.; Gadaleta-Caldarola, G.; Brandi, G. First-Line Immune Checkpoint Inhibitor-Based Combinations in Unresectable Hepatocellular Carcinoma: Current Management and Future Challenges. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 1245–1251.
- Rizzo, A.; Ricci, A.D. PD-L1, TMB, and Other Potential Predictors of Response to Immunotherapy for Hepatocellular Carcinoma: How Can They Assist Drug Clinical Trials? Expert Opin. Investig. Drugs 2022, 31, 415–423.
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC Strategy for Prognosis Prediction and Treatment Recommendation: The 2022 Update. J. Hepatol. 2022, 76, 681–693.
- Massarweh, N.N.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control 2017, 24, 1073274817729245.
- Zabron, A.; Edwards, R.J.; Khan, S.A. The Challenge of Cholangiocarcinoma: Dissecting the Molecular Mechanisms of an Insidious Cancer. Dis. Model. Mech. 2013, 6, 281–292.
- Krenzien, F.; Nevermann, N.; Krombholz, A.; Benzing, C.; Haber, P.; Fehrenbach, U.; Lurje, G.; Pelzer, U.; Pratschke, J.; Schmelzle, M.; et al. Treatment of Intrahepatic Cholangiocarcinoma-A Multidisciplinary Approach. Cancers 2022, 14, 362.
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next Horizon in Mechanisms and Management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588.
- Embade, N.; Fernández-Ramos, D.; Varela-Rey, M.; Beraza, N.; Sini, M.; de Juan, V.G.; Woodhoo, A.; Martínez-López, N.; Rodríguez-Iruretagoyena, B.; Bustamante, F.J.; et al. Mdm2 Regulates HuR Stability in Human Liver and Colon Cancer through Neddylation. Hepatology 2012, 55, 1237–1248.
- Zhu, H.; Berkova, Z.; Mathur, R.; Sehgal, L.; Khashab, T.; Tao, R.-H.; Ao, X.; Feng, L.; Sabichi, A.L.; Blechacz, B.; et al. HuR Suppresses Fas Expression and Correlates with Patient Outcome in Liver Cancer. Mol. Cancer Res. 2015, 13, 809–818.
- Vázquez–Chantada, M.; Fernández–Ramos, D.; Embade, N.; Martínez–Lopez, N.; Varela–Rey, M.; Woodhoo, A.; Luka, Z.; Wagner, C.; Anglim, P.P.; Finnell, R.H.; et al. HuR/Methyl-HuR and AUF1 Regulate the MAT Expressed During Liver Proliferation, Differentiation, and Carcinogenesis. Gastroenterology 2010, 138, 1943–1953.e3.
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e23.
- Toyota, K.; Murakami, Y.; Kondo, N.; Uemura, K.; Nakagawa, N.; Takahashi, S.; Sueda, T. Cytoplasmic Hu-Antigen R (HuR) Expression Is Associated with Poor Survival in Patients with Surgically Resected Cholangiocarcinoma Treated with Adjuvant Gemcitabine-Based Chemotherapy. Ann. Surg. Oncol. 2018, 25, 1202–1210.
- Abdelmohsen, K.; Gorospe, M. Posttranscriptional Regulation of Cancer Traits by HuR. Wiley Interdiscip. Rev. RNA 2010, 1, 214–229.
- Villanueva, A.; Chiang, D.Y.; Newell, P.; Peix, J.; Thung, S.; Alsinet, C.; Tovar, V.; Roayaie, S.; Minguez, B.; Sole, M.; et al. Pivotal Role of MTOR Signaling in Hepatocellular Carcinoma. Gastroenterology 2008, 135, 1972–1983.e11.
- Kang, M.-J.; Ryu, B.-K.; Lee, M.-G.; Han, J.; Lee, J.-H.; Ha, T.-K.; Byun, D.-S.; Chae, K.-S.; Lee, B.-H.; Chun, H.S.; et al. NF-KappaB Activates Transcription of the RNA-Binding Factor HuR, via PI3K-AKT Signaling, to Promote Gastric Tumorigenesis. Gastroenterology 2008, 135, 2030–2042.e3.
- Wang, L.; Ding, Q.; Zhao, L.; Pan, Y.; Song, Z.; Qin, Y.; Yan, X. Decreased BMP-7 and p-Smad1/5/8 Expression, and Increased Levels of Gremlin in Hepatocellular Carcinoma. Oncol. Lett. 2018, 16, 2113–2118.
- Hung, C.-M.; Huang, W.-C.; Pan, H.-L.; Chien, P.-H.; Lin, C.-W.; Chen, L.-C.; Chien, Y.-F.; Lin, C.-C.; Leow, K.-H.; Chen, W.-S.; et al. Hepatitis B Virus X Upregulates HuR Protein Level to Stabilize HER2 Expression in Hepatocellular Carcinoma Cells. Bio. Res. Int. 2014, 2014, 827415.
- Martínez–Chantar, M.L.; Vázquez–Chantada, M.; Garnacho, M.; Latasa, M.U.; Varela–Rey, M.; Dotor, J.; Santamaria, M.; Martínez–Cruz, L.A.; Parada, L.A.; Lu, S.C.; et al. S–Adenosylmethionine Regulates Cytoplasmic HuR Via AMP–Activated Kinase. Gastroenterology 2006, 131, 223–232.
- Ghosh, M.; Aguila, H.L.; Michaud, J.; Ai, Y.; Wu, M.-T.; Hemmes, A.; Ristimaki, A.; Guo, C.; Furneaux, H.; Hla, T. Essential Role of the RNA-Binding Protein HuR in Progenitor Cell Survival in Mice. J. Clin. Investig. 2009, 119, 3530–3543.
- Masuda, K.; Abdelmohsen, K.; Kim, M.M.; Srikantan, S.; Lee, E.K.; Tominaga, K.; Selimyan, R.; Martindale, J.L.; Yang, X.; Lehrmann, E.; et al. Global Dissociation of HuR-MRNA Complexes Promotes Cell Survival after Ionizing Radiation. EMBO J. 2011, 30, 1040–1053.
- Lai, K.K.Y.; Kweon, S.-M.; Chi, F.; Hwang, E.; Kabe, Y.; Higashiyama, R.; Qin, L.; Yan, R.; Wu, R.P.; Lai, K.; et al. Stearoyl-CoA Desaturase Promotes Liver Fibrosis and Tumor Development in Mice via a Wnt Positive-Signaling Loop by Stabilization of Low-Density Lipoprotein-Receptor-Related Proteins 5 and 6. Gastroenterology 2017, 152, 1477–1491.
- Chen, Y.; Peng, C.; Chen, J.; Chen, D.; Yang, B.; He, B.; Hu, W.; Zhang, Y.; Liu, H.; Dai, L.; et al. WTAP Facilitates Progression of Hepatocellular Carcinoma via M6A-HuR-Dependent Epigenetic Silencing of ETS1. Mol. Cancer 2019, 18, 127.
- Zhang, R.; Wang, J. HuR Stabilizes TFAM MRNA in an ATM/P38-Dependent Manner in Ionizing Irradiated Cancer Cells. Cancer Sci. 2018, 109, 2446–2457.
- Zhang, L.-F.; Lou, J.-T.; Lu, M.-H.; Gao, C.; Zhao, S.; Li, B.; Liang, S.; Li, Y.; Li, D.; Liu, M.-F. Suppression of MiR-199a Maturation by HuR Is Crucial for Hypoxia-Induced Glycolytic Switch in Hepatocellular Carcinoma. EMBO J. 2015, 34, 2671–2685.
- Cao, C.; Sun, J.; Zhang, D.; Guo, X.; Xie, L.; Li, X.; Wu, D.; Liu, L. The Long Intergenic Noncoding RNA UFC1, a Target of MicroRNA 34a, Interacts with the MRNA Stabilizing Protein HuR to Increase Levels of β-Catenin in HCC Cells. Gastroenterology 2015, 148, 415–426.e18.
- Huang, J.-F.; Jiang, H.-Y.; Cai, H.; Liu, Y.; Zhu, Y.-Q.; Lin, S.-S.; Hu, T.-T.; Wang, T.-T.; Yang, W.-J.; Xiao, B.; et al. Genome-Wide Screening Identifies Oncofetal LncRNA Ptn-Dt Promoting the Proliferation of Hepatocellular Carcinoma Cells by Regulating the Ptn Receptor. Oncogene 2019, 38, 3428–3445.
- He, X.; Zheng, Y.; Zhang, Y.; Gan, Y.; Zhou, Y.; Liang, H.; Wu, D.; Ge, W.; Deng, J.; Xu, X. Long Non-Coding RNA AK058003, as a Precursor of MiR-15a, Interacts with HuR to Inhibit the Expression of γ-Synuclein in Hepatocellular Carcinoma Cells. Oncotarget 2017, 8, 9451–9465.
- Liu, B.; Yang, G.; Wang, X.; Liu, J.; Lu, Z.; Wang, Q.; Xu, B.; Liu, Z.; Li, J. CircBACH1 (Hsa_circ_0061395) Promotes Hepatocellular Carcinoma Growth by Regulating P27 Repression via HuR. J. Cell. Physiol. 2020, 235, 6929–6941.
- Liu, H.; Lan, T.; Li, H.; Xu, L.; Chen, X.; Liao, H.; Chen, X.; Du, J.; Cai, Y.; Wang, J.; et al. Circular RNA CircDLC1 Inhibits MMP1-Mediated Liver Cancer Progression via Interaction with HuR. Theranostics 2021, 11, 1396–1411.
- Wang, Y.; Gao, R.; Li, J.; Tang, S.; Li, S.; Tong, Q.; Li, S. Downregulation of Hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int. J. Nanomed. 2021, 16, 2803–2818.
- Martínez-López, N.; Varela-Rey, M.; Fernández-Ramos, D.; Woodhoo, A.; Vázquez-Chantada, M.; Embade, N.; Espinosa-Hevia, L.; Bustamante, F.J.; Parada, L.A.; Rodriguez, M.S.; et al. Activation of LKB1-Akt Pathway Independent of Phosphoinositide 3-Kinase Plays a Critical Role in the Proliferation of Hepatocellular Carcinoma from Nonalcoholic Steatohepatitis. Hepatology 2010, 52, 1621–1631.
- Chen, T.; Dai, X.; Dai, J.; Ding, C.; Zhang, Z.; Lin, Z.; Hu, J.; Lu, M.; Wang, Z.; Qi, Y.; et al. AFP Promotes HCC Progression by Suppressing the HuR-Mediated Fas/FADD Apoptotic Pathway. Cell Death Dis. 2020, 11, 822.
- Ji, E.; Kim, C.; Kang, H.; Ahn, S.; Jung, M.; Hong, Y.; Tak, H.; Lee, S.; Kim, W.; Lee, E.K. RNA Binding Protein HuR Promotes Autophagosome Formation by Regulating Expression of Autophagy-Related Proteins 5, 12, and 16 in Human Hepatocellular Carcinoma Cells. Mol. Cell. Biol. 2019, 39, e00508-18.
- Papatheofani, V.; Levidou, G.; Sarantis, P.; Koustas, E.; Karamouzis, M.V.; Pergaris, A.; Kouraklis, G.; Theocharis, S. HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment. Biomedicines 2021, 9, 119.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Entry Collection:
Gastrointestinal Disease
Revisions:
2 times
(View History)
Update Date:
24 Jun 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




