Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Nagendra Kaushik | -- | 1886 | 2022-06-10 09:35:38 | | | |
| 2 | Camila Xu | Meta information modification | 1886 | 2022-06-10 09:50:21 | | | | |
| 3 | Camila Xu | Meta information modification | 1886 | 2022-06-10 09:51:10 | | | | |
| 4 | RAJAN ROLTA | + 200 word(s) | 2086 | 2022-06-10 10:29:08 | | | | |
| 5 | RAJAN ROLTA | Meta information modification | 2086 | 2022-06-10 10:29:56 | | | | |
| 6 | Camila Xu | -135 word(s) | 1951 | 2022-06-10 10:39:20 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kaushik, N.; Chugh, A.; Khurana, N.; , .; Rolta, R.; Kaushik, N.; Choi, E.H. Structure, Sequence and Immunopathology of SARS-CoV-2. Encyclopedia. Available online: https://encyclopedia.pub/entry/23909 (accessed on 08 February 2026).
Kaushik N, Chugh A, Khurana N, , Rolta R, Kaushik N, et al. Structure, Sequence and Immunopathology of SARS-CoV-2. Encyclopedia. Available at: https://encyclopedia.pub/entry/23909. Accessed February 08, 2026.
Kaushik, Nagendra, Ananya Chugh, Nimisha Khurana, , Rajan Rolta, Neha Kaushik, Eun Ha Choi. "Structure, Sequence and Immunopathology of SARS-CoV-2" Encyclopedia, https://encyclopedia.pub/entry/23909 (accessed February 08, 2026).
Kaushik, N., Chugh, A., Khurana, N., , ., Rolta, R., Kaushik, N., & Choi, E.H. (2022, June 10). Structure, Sequence and Immunopathology of SARS-CoV-2. In Encyclopedia. https://encyclopedia.pub/entry/23909
Kaushik, Nagendra, et al. "Structure, Sequence and Immunopathology of SARS-CoV-2." Encyclopedia. Web. 10 June, 2022.
Copy Citation
Since November 2019, SARS-CoV-2 has been a matter of global concern due to its rapid spread, the millions of deaths it caused, and repeated waves of infections. One after another, many variants of this novel virus have come into existence due to its constant mutability, specifically in the spike glycoprotein region. The tally for variants of concern (VOCs), which already include Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and Delta (B.1.617.2), has increased to five with the latest appearance of Omicron (B.1.1.529).
SARS-CoV-2
variants of concern
spike glycoprotein
1. Introduction
It all started when a media statement on several cases of viral pneumonia was reported by the Wuhan Municipal Health Commission, which was noticed by the World Health Organisation (WHO). This local outbreak of the novel coronavirus disease (COVID-19) spread like wildfire within a short period of time, crippling Europe and other continents, and, on 11 March 2020, COVID-19 was characterized as a pandemic by the WHO. It is caused by a novel human pathogen SARS-CoV-2 from the Coronaviridae family [1][2]. The global spread and high virulence of this virus has led to the appearance of many variants in various geographical locations. As stated by the WHO, some of these are emerging variants of concern (VOCs), which include Alpha (lineage B.1.1.7), Beta (lineage B.1.351), Gamma (lineage P.1), Delta (B.1.617.2), and Omicron (lineage B.1.1.529).
2. Structure and Sequence of SARS-CoV-2
Similar to SARS-CoVs and MERS-CoVs, which also belong to the Coronaviridae family, SARS-CoV-2 is an enveloped, positive single-strand RNA virus [3][4][5]. Genome analysis of SARS-CoV-2 has shown that it contains 14 open reading frames (ORFs), which encode 31 proteins [6]. ORF1 and ORF1ab constitute two-thirds of the known 14 ORFs and encode 16 non-structural proteins (nsp). The remaining one-third of ORFs encodes for eleven accessory proteins and four structural proteins, membrane (M), envelope (E), spike (S) and nucleocapsid (N) [7], as shown in Figure 1a. The eleven accessory proteins include ORF3a, ORF3b, ORF3c, ORF3d, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c and ORF10 [6].
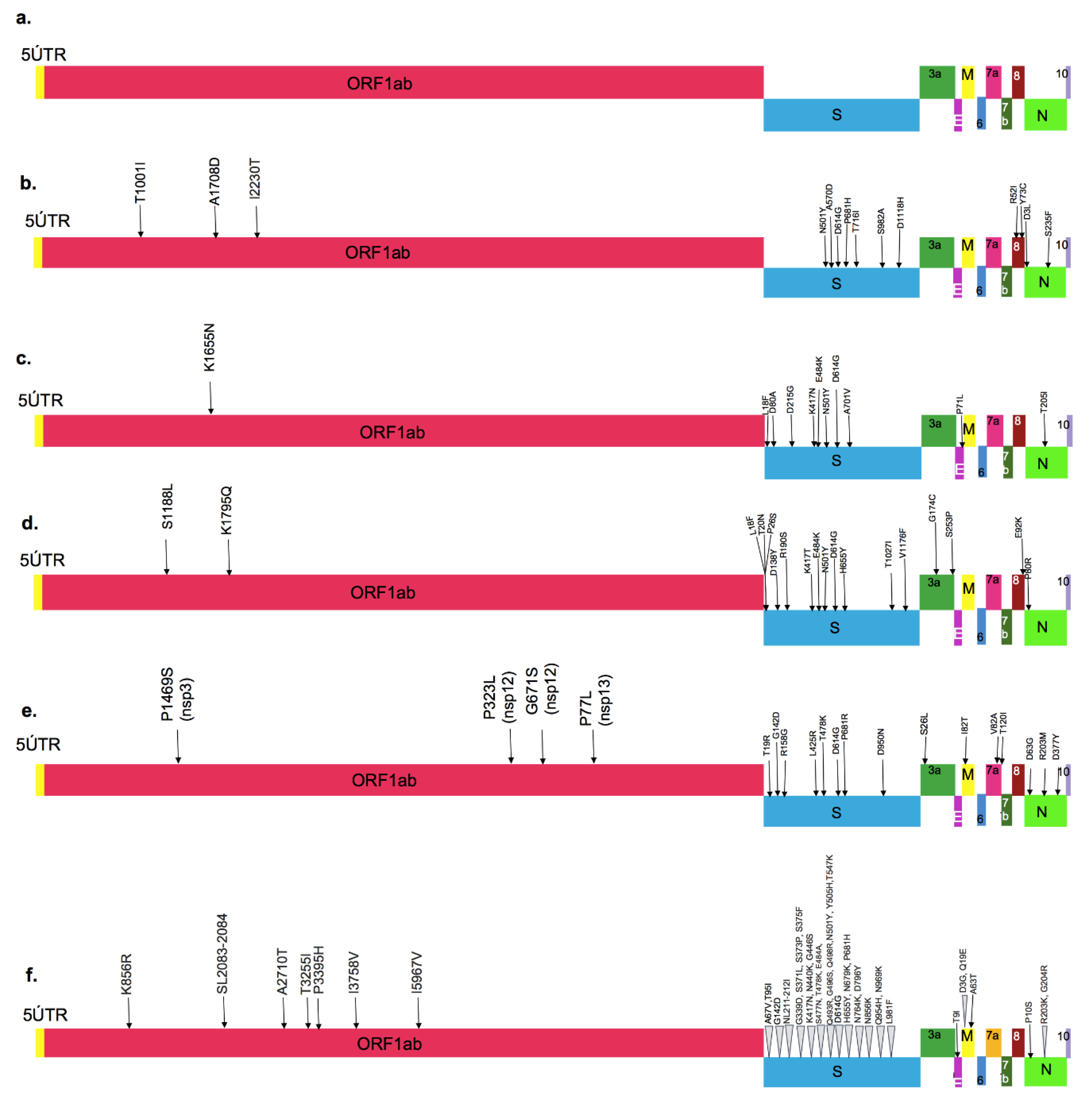 Figure 1. Major amino acid mutations marked on the entire SARS-CoV-2 sequence: (a) Wild-type strain, (b) Alpha variant, (c) Beta variant, (d) Gamma variant, (e) Delta variant and (f) Omicron variant. (M: membrane protein, E: envelope protein, S: Spike protein, N: Nucleocapsid protein, ORF: Open Reading Frame, UTR: Untranslated Region).
Figure 1. Major amino acid mutations marked on the entire SARS-CoV-2 sequence: (a) Wild-type strain, (b) Alpha variant, (c) Beta variant, (d) Gamma variant, (e) Delta variant and (f) Omicron variant. (M: membrane protein, E: envelope protein, S: Spike protein, N: Nucleocapsid protein, ORF: Open Reading Frame, UTR: Untranslated Region).The four structural proteins of SARS-CoV-2 and SARS-CoV have amino acid similarity above 90%, excluding the spike glycoprotein, where they diverge. The SARS-CoV-2 spike glycoprotein (1273 Aa) is 18 amino acids longer, compared to the S glycoprotein of SARS-CoV (1255 Aa) [8]. In fact, some of the mutations near the RBD region in the spike glycoprotein are known to make this virus more transmissible; therefore, this protein has been the focus of research to understand the emergence of new variants [9].
Spike glycoprotein is a club-shaped, trimeric class-I viral fusion protein with a length of ~20 nm. These spike glycoproteins form huge protrusions from the virus’s surface that facilitates virus entry into host cells [8][10][11]. It comprises two subunits: subunit S1 mediates receptor recognition, whereas S2 plays a role in membrane fusion [12]. The S1 subunit possesses a receptor-binding domain (RBD), which may be present either at the N-terminal domain (NTD) or C-terminal domain (CTD) or both [13]. The RBD of the virus mediates direct contact with human angiotensin-converting enzyme 2 (hACE2), which is the primary receptor for its entry into the host cell. The mutations in the RBD region of the spike protein are capable of forming closer contact with hACE2, responsible for a higher binding affinity and possibly/probably increased infectivity of VOCs [14]. The S2 subunit plays a role in the fusion of the virus membrane with the cell membrane of the host cell. In comparison to SARS-CoV, the spike glycoprotein of SARS-CoV-2 has four additional amino acid residues (PRRA) at the intersection of S1/S2, generating an S1/S2 protease cleavage site (PRRAR) [7][8] (Figure 2i and Figure 3). It is cleaved by cellular cathepsin L and transmembrane protease serine 2 (TMPRSS2), facilitating the virus entry through the plasma membrane or the endosomal membrane [7][15]. The spike glycoprotein can acquire two structural states: a closed state (Figure 2(iia)) and an open state (Figure 2(iib–d)) [16]. In a closed state, the RBDs are shielded by the N-terminal domains (NTDs) and the three recognition motifs of this trimeric protein do not protrude from the interface. In contrast, in an open state, the RBD is in the “up” conformation where one RBD is exposed upwards, away from the viral membrane, mediating the fusion of viral and host cell membranes (Figure 2(iib)) [16]. This open state is essential for SARS-CoV-2 and host cell membrane fusion, as this facilitates the passage of the virus into the host cells. Some mutations in the spike region promote an enhanced open conformation of the RBD [17].
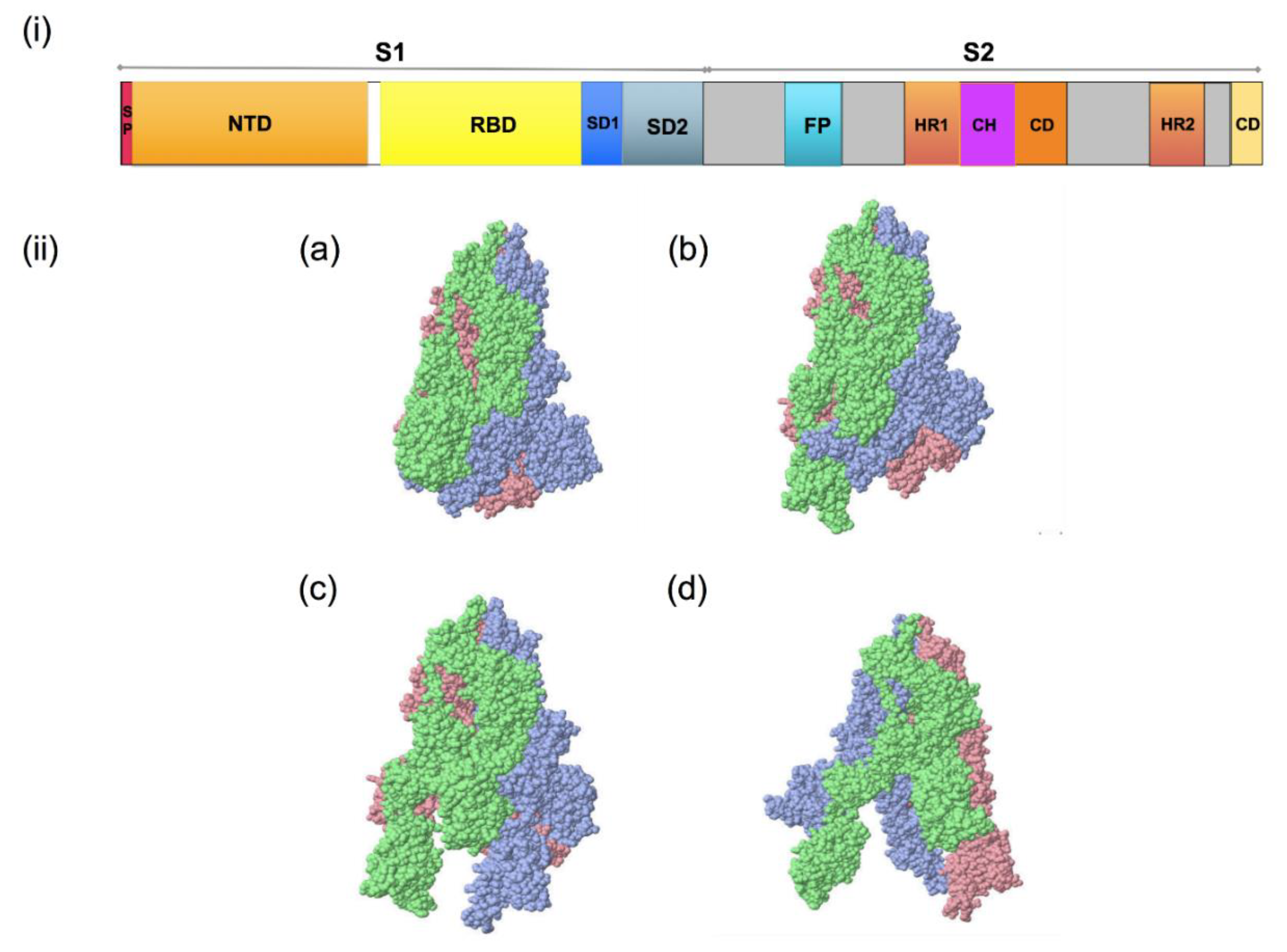 Figure 2. Spike glycoprotein of SARS-CoV-2; (i): S1/S2 protease cleavage site; SP: signal peptide; NTD: N-terminal domain; RBD: receptor-binding domain; RBM: receptor-binding motif; FP: fusion peptide; HR1: heptapeptide domain 1; HP2: heptapeptide domain 2; TM: transmembrane domain; CD: cytoplasmic domain 3. (ii): SARS-CoV-2 spike glycoprotein trimeric structure showing chains A (pink), B (blue) and C (green) of (a) closed state (7DF3), (b) 1 RBD up (7LWT), (c) 2 RBD up (7LYK) and (d) 3 RBD up (7CAK) (figure visualized using USF Chimera).
Figure 2. Spike glycoprotein of SARS-CoV-2; (i): S1/S2 protease cleavage site; SP: signal peptide; NTD: N-terminal domain; RBD: receptor-binding domain; RBM: receptor-binding motif; FP: fusion peptide; HR1: heptapeptide domain 1; HP2: heptapeptide domain 2; TM: transmembrane domain; CD: cytoplasmic domain 3. (ii): SARS-CoV-2 spike glycoprotein trimeric structure showing chains A (pink), B (blue) and C (green) of (a) closed state (7DF3), (b) 1 RBD up (7LWT), (c) 2 RBD up (7LYK) and (d) 3 RBD up (7CAK) (figure visualized using USF Chimera).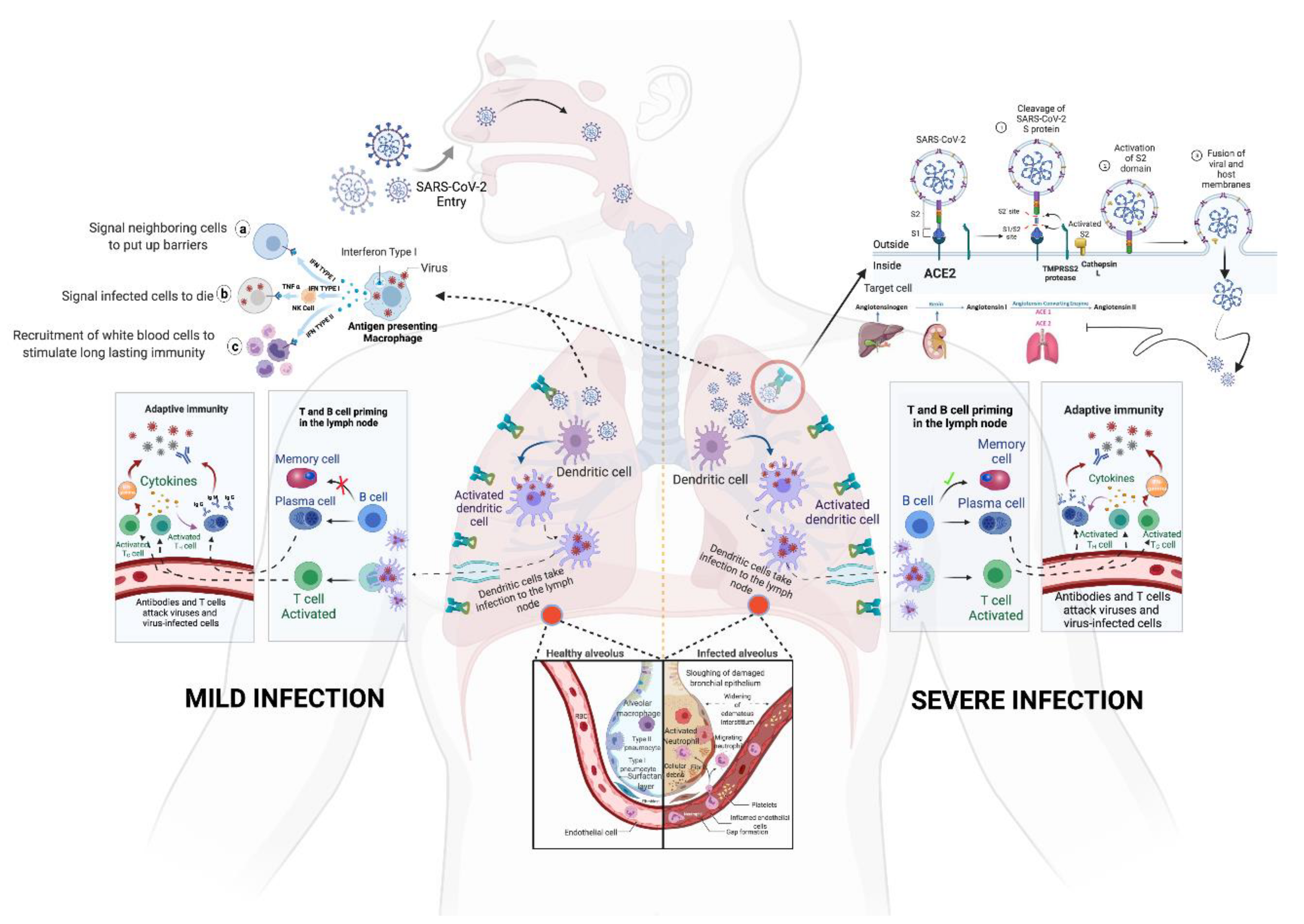 Figure 3. The entry and immunopathology of SARS-CoV-2 in the human body. Adapted from “Mechanism of SARS-CoV-2 Viral Entry, Acute Respiratory Distress Syndrome (ARDS), Adaptive Immunity and Interferons”, by biorender.com (accessed on 5 January 2022). Retrieved from https://app.biorender.com/biorender-templates; image for “memory cell and IFN gamma” from <https://reactome.org/PathwayBrowser/#/R-HSA-168256&SEL=R-HSA-1280218> and image for “ACE2” from https://reactome.org/PathwayBrowser/#/R-HSA-2022377&SEL=R-HSA-2022344&PATH=R-HSA-392499,R-HSA-2980736&FLG=Q9BYF1. Reactome, (all accessed on 30 December 2021).
Figure 3. The entry and immunopathology of SARS-CoV-2 in the human body. Adapted from “Mechanism of SARS-CoV-2 Viral Entry, Acute Respiratory Distress Syndrome (ARDS), Adaptive Immunity and Interferons”, by biorender.com (accessed on 5 January 2022). Retrieved from https://app.biorender.com/biorender-templates; image for “memory cell and IFN gamma” from <https://reactome.org/PathwayBrowser/#/R-HSA-168256&SEL=R-HSA-1280218> and image for “ACE2” from https://reactome.org/PathwayBrowser/#/R-HSA-2022377&SEL=R-HSA-2022344&PATH=R-HSA-392499,R-HSA-2980736&FLG=Q9BYF1. Reactome, (all accessed on 30 December 2021).Apart from the spike mutations, 13 different ORFs of the SARS-CoV-2 were also found to have mutations. A mutation in the ORF3a protein was observed to be linked to a higher level of infection and mortality rate due to infection caused by this virus [18]. The RNA-dependent RNA-polymerase of the SARS-CoV-2 has also been subjected to mutation located at the 14,408th position, which causes improper proofreading activity [19].
3. Immunopathology of SARS-CoV-2
Throughout the evolution of SARS-CoV-2, it has been observed that mutations and the response of the immune system towards those mutations have become the controlling factors of the pathogenicity of this virus. The host cell membrane has the protein receptor ACE-2, which influences the virus’s passage into the cell [20][21]. The virus enters the host cell by fusion of spike glycoprotein with the ACE-2 receptor on the cell membrane [22]. This induces conformational alteration in the plasma membrane and facilitates receptor-mediated endocytosis (Figure 3) [23]. Once uncoating is performed inside the cell, all proteins are removed and viral RNA is released into the cytoplasm. Then, spike glycoproteins inhibit ACE-2 and downregulate it. The downregulation of ACE-2 leads to the compensatory increase in ACE-1 and causes the excessive production of angiotensin-II, consequently enhancing pulmonary vascular permeability [24][25].
Once the cell is infected by the virus, it stimulates the production of interferon α in infected cells, which alerts the adjacent cells to stimulate the antiviral program. This leads to the production of interferon γ in the immune cells, which, in turn, triggers the immune system to destroy the infected cells. Virus-infected cells lose surface MHC I expression, which leads to a reduced inhibitory signal in NK cells. Additionally, cellular stress due to the viral infection upregulates the activating receptors of NK cells leading to the elimination of infected cells. Simultaneously, cytotoxic T cells are alerted when a fragment of viral protein is transported onto the cell’s surface by MHC I, which presents antigens from the virus within the cell [24]. Dendritic cells present antigens on their surface, which interacts with T cells and B cells to produce antigen-specific antibodies (Figure 3). Additionally, phagocytes play a significant role in innate immunity by ingesting the free virus particles and contribute to adaptive immunity by presenting antigens to lymphocytes. Simultaneously, certain chemokines, such as CXCL10 and CCL2, are released in order to guide immune cells towards the infected region and, hence, destroy the infected cells [26][27]. Studies report that the failure of switching between innate to adaptive responses disables the formation of memory cells, which increases the risk of recurring infections. Alluding to the above mechanism, the human body is well designed to protect itself against pathogens. Yet, SARS-CoV-2 successfully escapes and disrupts the immune response by generating various viral proteins, such as 3b and M, which inhibits the natural processes of the cell. One such protein, namely, PLpro, binds with the NF-κβ transcription factor (which is an important response generator of pro-inflammatory cytokines) and downregulates it. This leads to a reduced NF-κβ response and decreased production of pro-inflammatory cytokines [28], thus disabling the timely destruction of infected cells and leads to symptoms. Although the symptoms of infection by variants of SARS-CoV-2 are similar, their transmissibility varies due to mutations primarily in the spike glycoprotein [9][29].
The genomic sequencing of the virus from various locations at a larger scale has played a vital role in tracing the mutations (Figure 4). Evidence shows that some of these mutations, such as K417N and E484K, elude the immune system, i.e., they can escape the neutralizing antibodies, even after a person is vaccinated (Table 1) [30][31]. This integration of genome sequencing on a global scale, with routine sanitation practices and good quality clinical management, can equip the world against any upcoming crisis of this type. Therefore, it is important to consider the mutations currently known, and predict emerging prospective mutations that should be considered when testing and developing vaccines.
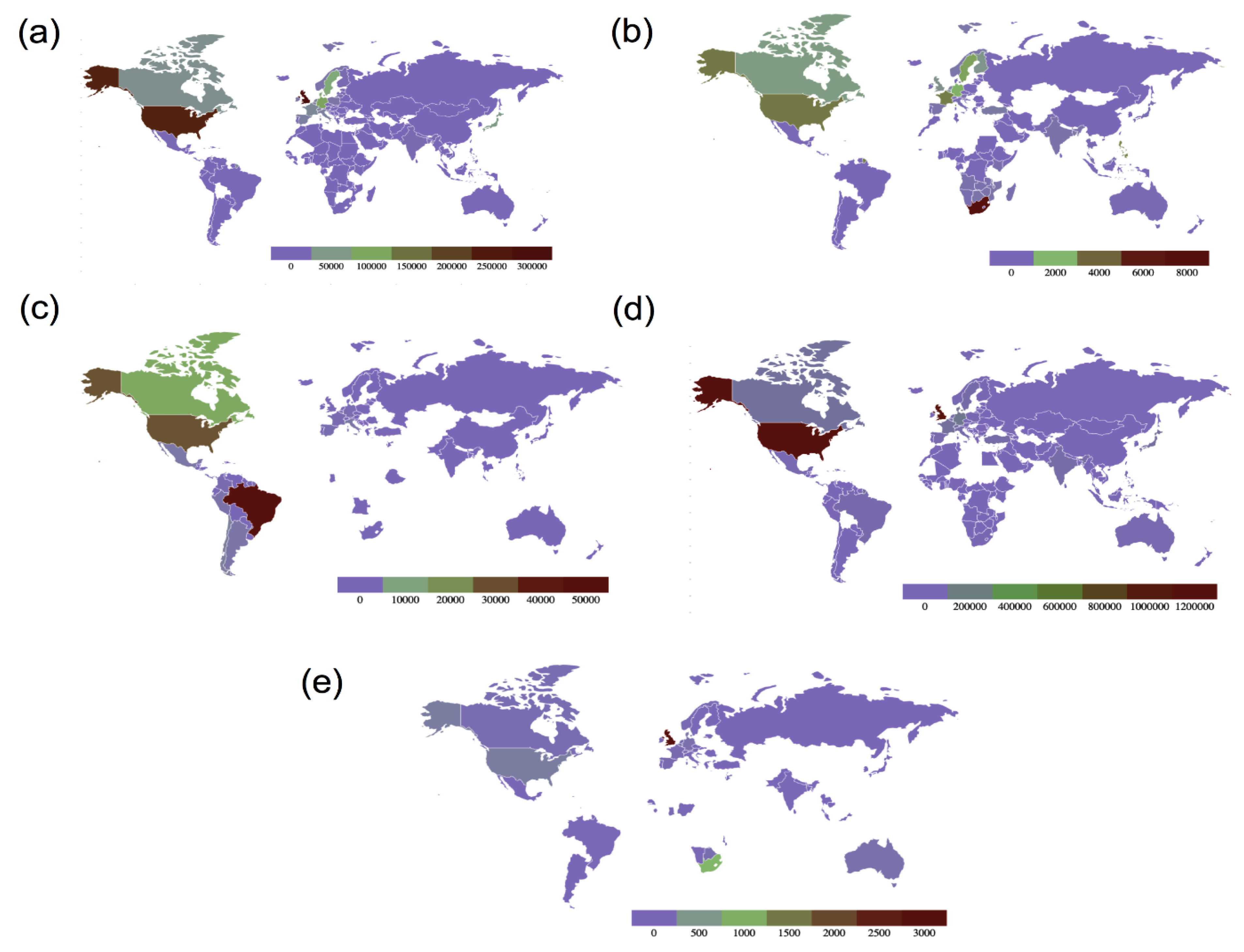 Figure 4. Maps showing global country-wise sequence count of all variants of concern as of 13 September 2021. (a) Sequence count of B.1.1.7 (Alpha variant), (b) sequence count of B.1.351 (Beta variant), (c) sequence count of P.1 (Gamma variant), (d) Sequence count of B.1.617.2 (Delta variant) and (e) sequence count of B.1.1.529 (Omicron variant) (data source: GISAID, Figure redrawn using GeoMaps).
Figure 4. Maps showing global country-wise sequence count of all variants of concern as of 13 September 2021. (a) Sequence count of B.1.1.7 (Alpha variant), (b) sequence count of B.1.351 (Beta variant), (c) sequence count of P.1 (Gamma variant), (d) Sequence count of B.1.617.2 (Delta variant) and (e) sequence count of B.1.1.529 (Omicron variant) (data source: GISAID, Figure redrawn using GeoMaps).Table 1. SARS-CoV-2 variants of concern (VOCs) with their date and place of emergence and important spike mutations, along with their effect on the variant’s overall transmissibility.
| WHO Label | Pango Lineage | Potential Emergence | Number of Countries Affected | Important Spike Mutations | Sequenced Genomes (GISAID) |
Impact of Mutations |
Transmissibility of Variant |
|---|---|---|---|---|---|---|---|
| Alpha | B.1.1.7 | UK, September 2020 | 170 | N501Y P681H |
11,49,443 | P681H: Slightly increases the S1–S2 cleavage [32]. N501Y: Enhances binding affinity between spike RBD domain and hACE2 receptor [33]. E484K *, L452R * |
~70% increase with respect to wild type |
| Beta | B.1.351 | South Africa, May 2020 |
112 | K417N E484K N501Y |
39,604 | K417N: Responsible for antibody-escape property of the virus [34]. N501Y: Same as above. E484K: Reduces antibody neutralisation and is responsible for increased infectivity [35]. |
20–113% increase with respect to wild type |
| Gamma | P.1 | Brazil, November 2020 | 80 | K417T E484K N501Y |
1,17,675 | K417T: Responsible for antibody escape property of the virus [34]. N501Y: Same as above. E484K: Same as above. |
~161% increase with respect to wild type |
| Delta | B.1.617.2 | India, October 2020 |
105 | L452R T478K P681R |
34,37,588 | T478K: Helps evade neutralising antibodies. Enhances viral infectivity [36]. L452R: Impairs antibody neutralization. Increases viral infectivity [34][37]. P681R: Increases the rate of S1–S2 cleavage, and hence results in enhanced transmissibility [35]. |
|
| Omicron | B.1.1.529 | Multiple countries, November 2021 |
- | H655Y N679K P681H E484A Q498R N501Y |
5352 | N501Y: Same as above. H655Y: May be associated with increased transmissibility as it is proximal to the Furin cleavage site, hence aids in increased spike cleavage (CoVariants-GISAID). N679K: Associated with increased transmissibility (CoVariants-GISAID). P681H: Slightly increases the S1–S2 cleavage [32]. E484A: May be associated with immune escape (CoVariants-GISAID). Q498R: Enhances binding affinity with ACE2 receptor in combination with N501Y (CoVariants-GISAID). |
- |
References
- Li, C.; Yang, Y.; Ren, L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect. Genet. Evol. 2020, 82, 104285.
- Phartyal, R.; Verma, M. SARS-CoV-2 Mutations: An Insight. In Human Viruses: Diseases, Treatments and Vaccines—The New Insights; Ahmad, S.I., Ed.; Springer: Cham, Switzerland, 2021; pp. 551–563. ISBN 978-3-030-71165-8.
- Gupta, V.; Haider, S.; Verma, M.; Singhvi, N.; Ponnusamy, K.; Malik, M.Z.; Verma, H.; Kumar, R.; Sood, U.; Hira, P.; et al. Comparative Genomics and Integrated Network Approach Unveiled Undirected Phylogeny Patterns, Co-mutational Hot Spots, Functional Cross Talk, and Regulatory Interactions in SARS-CoV-2. MSystems 2021, 6, e00030-21.
- Kumar, R.; Verma, H.; Singhvi, N.; Sood, U.; Gupta, V.; Singh, M.; Kumari, R.; Hira, P.; Nagar, S.; Talwar, C.; et al. Comparative Genomic Analysis of Rapidly Evolving SARS-CoV-2 Reveals Mosaic Pattern of Phylogeographical Distribution. MSystems 2021, 5, e00505-20.
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.-Y.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020, 5, 1408–1417.
- Redondo, N.; Zaldívar-López, S.; Garrido, J.J.; Montoya, M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 2021, 12, 708264.
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115.
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154.
- Soh, S.M.; Kim, Y.; Kim, C.; Jang, U.S.; Lee, H.-R. The rapid adaptation of SARS-CoV-2–rise of the variants: Transmission and resistance. J. Microbiol. 2021, 59, 807–818.
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192.
- Hasöksüz, M.; Kiliç, S.; Saraç, F. Coronaviruses and SARS-COV-2. Turk. J. Med. Sci. 2020, 50, 549–556.
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734.
- Turoňová, B.; Sikora, M.; Schürmann, C.; Hagen, W.J.H.; Welsch, S.; Blanc, F.E.C.; Von Bülow, S.; Gecht, M.; Bagola, K.; Hörner, C.; et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 2020, 370, 203–208.
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149.
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792.
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.
- Gobeil, S.M.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Saunders, K.; Edwards, R.J.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 2021, 373, eabi6226.
- Majumdar, P.; Niyogi, S. ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020, 148, e262.
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 179.
- Wei, J.; Alfajaro, M.M.; DeWeirdt, P.C.; Hanna, R.E.; Lu-Culligan, W.J.; Cai, W.L.; Strine, M.S.; Zhang, S.-M.; Graziano, V.R.; Schmitz, C.O.; et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 2021, 184, 76–91.e13.
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687.
- Liang, Y.; Wang, M.-L.; Chien, C.-S.; Yarmishyn, A.A.; Yang, Y.-P.; Lai, W.-Y.; Luo, Y.-H.; Lin, Y.-T.; Chen, Y.-J.; Chang, P.-C.; et al. Highlight of Immune Pathogenic Response and Hematopathologic Effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 Infection. Front. Immunol. 2020, 11, 1022.
- Daniloski, Z.; Jordan, T.X.; Wessels, H.-H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105.e16.
- Torbati, E.; Krause, K.L.; Ussher, J.E. The Immune Response to SARS-CoV-2 and Variants of Concern. Viruses 2021, 13, 1911.
- Cheng, Y.-R.; Li, X.; Zhao, X.; Lin, H. Cell Entry of Animal Coronaviruses. Viruses 2021, 13, 1977.
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past. Front. Immunol. 2020, 11, 1949.
- Catanzaro, M.; Fagiani, F.; Racchi, M.; Corsini, E.; Govoni, S.; Lanni, C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5, 84.
- De Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534.
- Graham, M.S.; Sudre, C.H.; May, A.; Antonelli, M.; Murray, B.; Varsavsky, T.; Kläser, K.; Canas, L.S.; Molteni, E.; Modat, M.; et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: An ecological study. Lancet Public Health 2021, 6, e335–e345.
- Mehra, R.; Kepp, K.P. Structure and Mutations of SARS-CoV-2 Spike Protein: A Focused Overview. ACS Infect. Dis. 2022, 8, 29–58.
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424.
- Winger, A.; Caspari, T. The Spike of Concern—The Novel Variants of SARS-CoV-2. Viruses 2021, 13, 1002.
- Luan, B.; Wang, H.; Huynh, T. Molecular Mechanism of the N501Y Mutation for Enhanced Binding between SARS-CoV-2’s Spike Protein and Human ACE2 Receptor. BioRxiv 2021, BioRxiv:2021.01.04.425316.
- Chakraborti, P.K.; Matange, N.; Nandicoori, V.K.; Singh, Y.; Tyagi, J.S.; Visweswariah, S.S. Signalling mechanisms in Mycobacteria. Tuberculosis 2011, 91, 432–440.
- Barton, M.I.; MacGowan, S.A.; Kutuzov, M.A.; Dushek, O.; Barton, G.J.; van der Merwe, P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. eLife 2021, 10, e70658.
- Di Giacomo, S.; Mercatelli, D.; Rakhimov, A.; Giorgi, F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021, 93, 5638–5643.
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. Convergent Evolution of SARS-CoV-2 Spike Mutations, L452R, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. BioRxiv 2021, BioRxiv:2021.04.22.440932.
- Teo, S.P. Review of COVID-19 mRNA Vaccines: BNT162b2 and mRNA-1273. J. Pharm. Pract. 2021, 8971900211009650.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Entry Collection:
COVID-19
Revisions:
6 times
(View History)
Update Date:
10 Jun 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




