Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sandra Marcia Muxel | -- | 1403 | 2022-05-12 14:19:38 | | | |
| 2 | Catherine Yang | + 1 word(s) | 1404 | 2022-05-13 03:30:19 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Muxel, S.M.; Maia Acuña, S. MicroRNAs. Encyclopedia. Available online: https://encyclopedia.pub/entry/22873 (accessed on 07 February 2026).
Muxel SM, Maia Acuña S. MicroRNAs. Encyclopedia. Available at: https://encyclopedia.pub/entry/22873. Accessed February 07, 2026.
Muxel, Sandra Marcia, Stephanie Maia Acuña. "MicroRNAs" Encyclopedia, https://encyclopedia.pub/entry/22873 (accessed February 07, 2026).
Muxel, S.M., & Maia Acuña, S. (2022, May 12). MicroRNAs. In Encyclopedia. https://encyclopedia.pub/entry/22873
Muxel, Sandra Marcia and Stephanie Maia Acuña. "MicroRNAs." Encyclopedia. Web. 12 May, 2022.
Copy Citation
In the past decade, microRNAs (miRNAs), a group of noncoding small RNAs, have emerged as functionally significant regulatory molecules with the significant capability of fine-tuning biological processes. The important role of miRNAs in inflammation and immune responses is highlighted by studies in which the regulation of miRNAs in the host was shown to be related to infectious diseases and associated with the eradication or susceptibility of the infection.
microRNAs
gene expression
post-transcriptional
1. Introduction
The discovery of noncoding RNAs (ncRNAs) has revolutionized the field of molecular biology. These ncRNAs do not code for proteins but globally impact genome maintenance and gene expression [1][2][3][4]. These RNAs can be categorized according to their length, localization, and function, such as ncRNAs that regulate gene expression, as long noncoding RNAs (lncRNAs), microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs); RNA maturation, as small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs); and protein synthesis, as ribosomal RNA (rRNAs) and transfer RNAs (tRNAs) [5][6].
miRNAs (typically 21 nucleotides) are a class of ncRNAs which interact with the 3′ untranslated region (3′ UTR) of messenger RNAs (mRNAs), leading to mRNA degradation or translational repression [7][8][9][10]. The involvement of miRNAs in a variety of biological processes, such as chronic pathologies and infectious diseases, illustrates their complexity as they have been described as being able to simultaneously interact with diverse molecules, such as RNA binding proteins (RBPs), making the miRNA recognition site more accessible to the RNA-induced silencing complex (RISC), which also enables the processing of pri-miRNAs [11][12] or lncRNAs [1][13], controlling different points of the gene expression flux.
Host–pathogen interactions result in signaling and physiological modifications in host cells that induce the miRNA-mediated post-transcriptional regulation of genes involved in the inflammatory response during the induction of the immune response [14][15]. These miRNAs are involved in the modulation of both innate and adaptive immune responses [16]. In recent years, the alteration of miRNA expression has been studied extensively in cancer or infectious diseases caused by bacteria, viruses, and parasites [17][18][19][20].
The proposition of miRNAs as potentially non-invasive biomarkers for clinical diagnosis of diseases, using patient plasma, has prompted descriptions of tumor-derived miRNAs in that medium [21][22]. Also, some studies have shown that miRNAs are differentially expressed in patients with hepatotoxicity [23], artery disease [24], and infectious diseases [19][20][25].
2. miRNAs—Biogenesis and Gene Expression Regulation
miRNAs are transcribed from intergenic regions as either monocistronic or polycistronic (miRNA-clusters) transcripts [26][27][28]. Those sequences can have their own promoter region or can depend on the transcription of host genes if they are intragenic [26][27][28]. They can be encoded in exonic or intronic regions and transcribed in the same direction as that of the pre-messenger RNA, leading to the use of the promoter region of mRNAs for their transcription [29] (Figure 1). miRNAs are transcribed by RNA polymerase II and fold into long double-strand primary miRNA transcripts (pri-miRNA) [7]. In the nucleus, the class 2 RNase III DROSHA and DGCR8 (a double-strand RNA-binding protein, also known as Pasha) complex recognizes features in the hairpin structures of pri-miRNA and processes the molecule to form the precursor miRNA transcript (pre-miRNA) [30][31]. The pre-miRNA is coupled with the Exportin 5 protein and exported to the cytoplasm [32] where an RNase III family protein Dicer complexed with TRBP (transactivation response element RNA-binding protein) recognizes and processes the pre-miRNA into the miRNA-duplex, a mature miRNA [33][34]. The functional strand of the mature miRNA is loaded into the RISC, coupled with the argonaute (AGO) protein family as important components of ribonucleoprotein (RNP) complexes (miRNPs) which guide interactions with the target mRNA, leading to the regulation of gene expression [33][35][36][37].
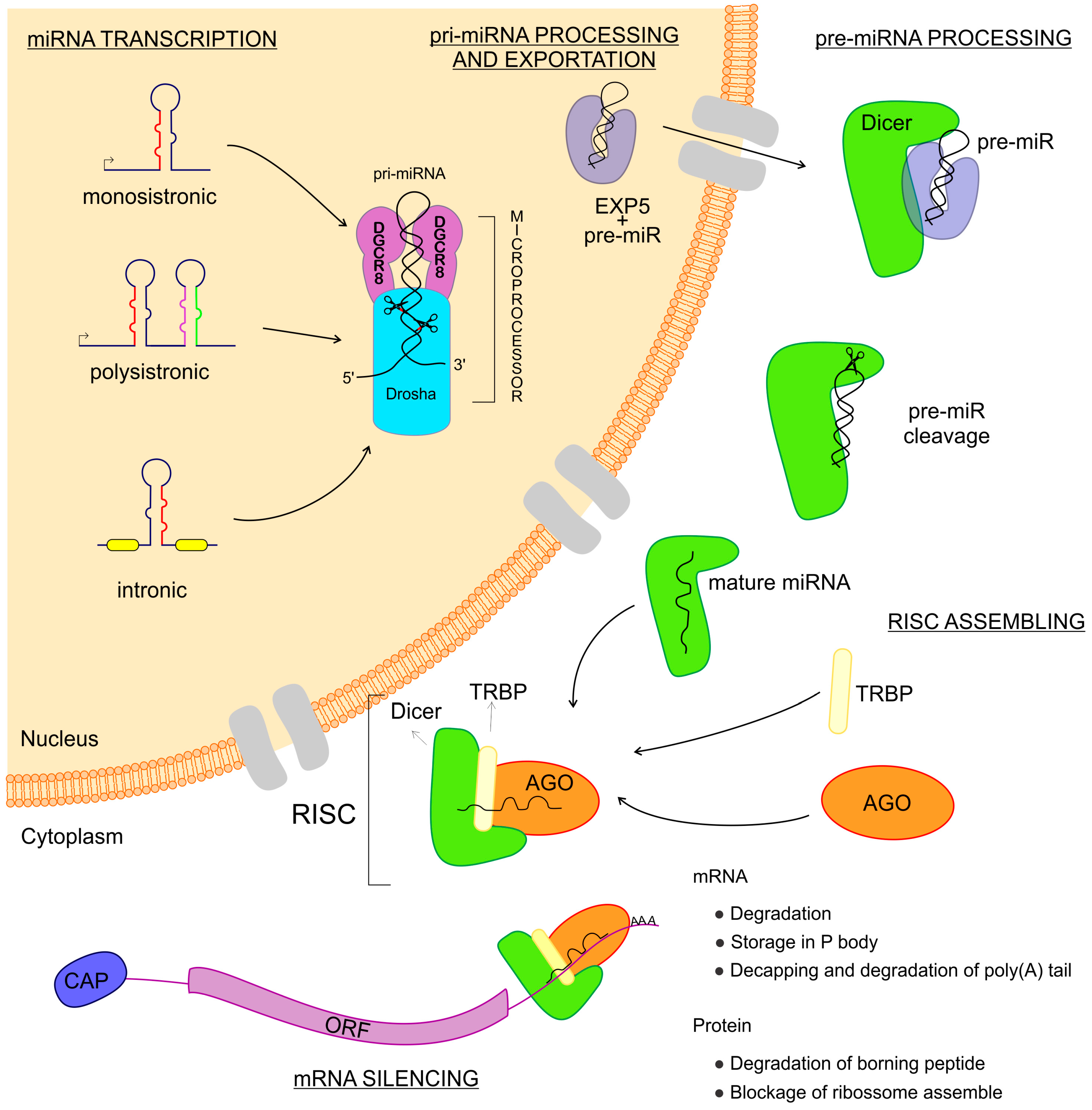
Figure 1. Biogenesis of microRNAs (miRNAs). The miRNAs are small non-coding RNAs transcribed from DNA sequences which can be monocistronic or polycistronic, comprised within exons, introns, or a unique host gene. Right after the transcription, this new RNA sequence is called primary-miRNA (pri-miRNA), which is folded in a hairpin conformation and coupled with the microprocessor: a combination of DGCR8 and Drosha RNases which cut the pri-miRNA, making it a pre-miRNA. Afterward, the pre-miRNA is coupled to Exportin 5 and then exported to the cytoplasm where this complex is found by Dicer, which cuts the pre-miRNA, releasing two mature miRNA arms. The mature miRNA complexed to the Dicer can now couple with the Argonaute and TRBP proteins, thus assembling the RNA Induced Silencing Complex–RISC. When the RISC is done, it can find the messenger RNAs that are the targets to the miRNA complexed. Once found, the message is now silenced and the gene expression regulated.
miRNAs regulate gene expression at a post-transcriptional level in a sequence-specific manner, exerting a massive biological impact [7]. Research has been dedicated toward attempting to understand the mechanisms of post-transcriptional regulation mediated by miRNAs, with studies performed in vitro, in vivo, and in cell-free extracts, as well as using bioinformatics prediction tools to demonstrate the putative regulation, by miRNAs, of nearly 30% of all protein-coding genes in mammalian cells [38]. miRNAs can regulate translation by (a) repressing the initiation of protein translation of 7-methylguanosine (m7GpppN)-capped mRNAs, inhibiting the binding of the eukaryotic translation initiation factor (eIF) subunits eIF4E, eIF4F, and eIF4G, which promote the scaffolding of mRNA for association of the ribosome initiation complex [39][40][41]; (b) preventing the association of ribosome 60 S subunit with 40 S and mRNA via interference of the binding between eIF6 and 60 S, which can be mediated by the AGO2–Dicer–TRBP complex [42]; and (c) blocking elongation via the miRNA–mRNA association with active polysomes, impacting the post-initiation step of translation [43]. miRNAs can also regulate mRNA destabilization by recruiting decay machinery components, leading to mRNA poly(A) tail deadenylation by exonuclease activity in 3′→5′ degradation, or decapping followed by 5′→3′ exonuclease activity [44][45]. In addition, miRNAs are secreted into vesicles, as exosomes, or into extracellular fluids and circulation, highlighting the potential roles of extracellular miRNAs in mediating intercellular communications and as biomarkers for a variety of diseases, including infectious diseases [46].
2. Novel Perspectives: MicroRNAs as Diagnostic Markers and Therapeutic Targets
miRNAs have been emerging as biomarkers and treatment assets of many diseases [47][48]. Why not think about them as a profitable approach to better diagnose and properly treat parasitic diseases? Here, the researchers revised how miRNAs are related to parasitic diseases and included not only the vertebrate hosts, but also the invertebrate ones. Moreover, parasitic diseases affect animals that provide economical and emotional benefits to people. It is inarguable that the loss of support pets or livestock animals causes socio-economic and psychological consequences for people. Nevertheless, a deep gap persists in the development of therapeutic strategies for neglected diseases.
Although, there is an increasing number of studies that are being done in miRNA, however translational research of miRNA still remains a challenge. Earlier this year, Hanna et al. reviewed the role of miRNAs in clinical studies. They observed that findings in miRNA research are limited to academia, often do not make it through phase 3 or 4 of clinical trials, and there is a gap between basic science and clinical application [47][48].
Regarding the neglected diseases, researchers have dedicated decades to the development of new drugs and identifying new biomarkers of disease progression. Despite this, the use of miRNAs as a biomarker and its potential for treatment are poorly explored when compared with hotspot diseases like cancer, cardiac and circulatory diseases, and neurological disorders. Currently, there are only two clinical trials regarding parasitosis and both are cohort studies. The first one was developed in Chicago city, USA, correlating the miRNA profile with the protein expression in toxoplasmosis patients [49]. The other study is focused on figuring out what miRNAs are modified in plasma samples from patients who suffer from acute myocarditis caused by the Chagas disease from Salvador city in Brazil. This study is available in the clinical trials database to consult (<https://clinicaltrials.gov/ct2/show/NCT01842880?term=miRNA&cond=Trypanosomiasis&draw=2&rank=1>).
Thus, the myriad of processes regulated by miRNAs makes them a very profitable approach to be studied, not only as biomarkers, but also as drugs in the treatment of pathologies [48]. Diversity of cell culture platforms, animal models, and the evaluation of circulating fluids (saliva, plasma, and serum) are now available to assess the treatment mechanisms, toxicity, and potential therapeutic efficacy of miRNA candidates in vitro [47][48]. Also, miRNAs can be used in the association of conventional treatments to target a large number of genes and pathways that deliver the host susceptibility to disease and ameliorate the prognosis. Further, the impact of specific miRNAs or a combination of some miRNAs on the regulation of immune responses in the hosts remains a significant challenge.
References
- Fernandes, J.C.R.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long non-coding RNAs in the regulation of gene expression: Physiology and disease. Non-Coding RNA 2019, 5, 17.
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110.
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The gencode v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789.
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108.
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46.
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166.
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Bagga, S.; Bracht, J.; Hunter, S.; Massirer, K.; Holtz, J.; Eachus, R.; Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005, 122, 553–563.
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773.
- Ben-Hamo, R.; Efroni, S. MicroRNA regulation of molecular pathways as a generic mechanism and as a core disease phenotype. Oncotarget 2015, 6, 1594–1604.
- Nussbacher, J.K.; Yeo, G.W. Systematic discovery of RNA binding proteins that regulate microRNA levels. Mol. Cell 2018, 69, 1005–1016.
- Hao, J.; Duan, F.F.; Wang, Y. MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 95–103.
- Yu, Y.; Nangia-Makker, P.; Farhana, L.; Majumdar, A.P.N. A novel mechanism of lncRNA and miRNA interaction: Ccat2 regulates mir-145 expression by suppressing its maturation process in colon cancer cells. Mol. Cancer 2017, 16, 155.
- Baltimore, D.; Boldin, M.P.; O’Connell, R.M.; Rao, D.S.; Taganov, K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845.
- O’Neill, L.A.; Sheedy, F.J.; McCoy, C.E. MicroRNAs: The fine-tuners of toll-like receptor signalling. Nat. Rev. Immunol 2011, 11, 163–175.
- Labbaye, C.; Testa, U. The emerging role of mir-146a in the control of hematopoiesis, immune function and cancer. J. Hematol. Oncol. 2012, 5, 13.
- Manzano-Roman, R.; Siles-Lucas, M. MicroRNAs in parasitic diseases: Potential for diagnosis and targeting. Mol. Biochem. Parasitol. 2012, 186, 81–86.
- Petrini, E.; Caviglia, G.P.; Abate, M.L.; Fagoonee, S.; Smedile, A.; Pellicano, R. MicroRNAs in hbv-related hepatocellular carcinoma: Functions and potential clinical applications. Panminerva Med. 2015, 57, 201–209.
- Reynoso, R.; Laufer, N.; Hackl, M.; Skalicky, S.; Monteforte, R.; Turk, G.; Carobene, M.; Quarleri, J.; Cahn, P.; Werner, R.; et al. MicroRNAs differentially present in the plasma of hiv elite controllers reduce hiv infection in vitro. Sci. Rep. 2014, 4, 5915.
- Fu, Y.; Yi, Z.; Wu, X.; Li, J.; Xu, F. Circulating microRNAs in patients with active pulmonary tuberculosis. J. Clin. Microbiol. 2011, 49, 4246–4251.
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518.
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large b-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675.
- Ozer, J.; Ratner, M.; Shaw, M.; Bailey, W.; Schomaker, S. The current state of serum biomarkers of hepatotoxicity. Toxicology 2008, 245, 194–205.
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Roxe, T.; Muller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684.
- Abd-El-Fattah, A.A.; Sadik, N.A.; Shaker, O.G.; Aboulftouh, M.L. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem. Biophys. 2013, 67, 875–884.
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20.
- Lai, F.; Gardini, A.; Zhang, A.; Shiekhattar, R. Integrator mediates the biogenesis of enhancer RNAs. Nature 2015, 525, 399–403.
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739.
- Lai, E.C.; Tomancak, P.; Williams, R.W.; Rubin, G.M. Computational identification of drosophila microRNA genes. Genome Biol. 2003, 4, R42.
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNAse iii drosha initiates microRNA processing. Nature 2003, 425, 415–419.
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.K.; Yeom, K.H.; Yang, W.Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between drosha and dgcr8. Cell 2009, 136, 75–84.
- Yamazawa, R.; Jiko, C.; Choi, S.; Park, I.Y.; Nakagawa, A.; Yamashita, E.; Lee, S.J. Structural basis for selective binding of export cargoes by exportin-5. Structure 2018, 26, 1393–1398 e1392.
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366.
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838.
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208.
- Wang, B.; Li, S.; Qi, H.H.; Chowdhury, D.; Shi, Y.; Novina, C.D. Distinct passenger strand and mRNA cleavage activities of human argonaute proteins. Nat. Struct. Mol. Biol. 2009, 16, 1259–1266.
- Vaucheret, H.; Vazquez, F.; Crete, P.; Bartel, D.P. The action of argonaute1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197.
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114.
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by let-7 microRNA in human cells. Science 2005, 309, 1573–1576.
- Humphreys, D.T.; Westman, B.J.; Martin, D.I.; Preiss, T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4e/cap and poly(a) tail function. Proc. Natl. Acad. Sci. USA 2005, 102, 16961–16966.
- Richter, J.D.; Sonenberg, N. Regulation of cap-dependent translation by eif4e inhibitory proteins. Nature 2005, 433, 477–480.
- Chendrimada, T.P.; Finn, K.J.; Ji, X.; Baillat, D.; Gregory, R.I.; Liebhaber, S.A.; Pasquinelli, A.E.; Shiekhattar, R. MicroRNA silencing through risc recruitment of eif6. Nature 2007, 447, 823–828.
- Maroney, P.A.; Yu, Y.; Fisher, J.; Nilsen, T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006, 13, 1102–1107.
- Parker, R.; Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127.
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22.
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49.
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143.
- Hewson, C.; Capraro, D.; Burdach, J.; Whitaker, N.; Morris, K.V. Extracellular vesicle associated long non-coding RNAs functionally enhance cell viability. Non-Coding RNA Res. 2016, 1, 3–11.
- Ngo, H.M.; Zhou, Y.; Lorenzi, H.; Wang, K.; Kim, T.K.; Zhou, Y.; El Bissati, K.; Mui, E.; Fraczek, L.; Rajagopala, S.V.; et al. Toxoplasma modulates signature pathways of human epilepsy, neurodegeneration & cancer. Sci. Rep. 2017, 7, 11496.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
970
Revisions:
2 times
(View History)
Update Date:
13 May 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




