Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Junlong Song | -- | 2006 | 2022-05-07 08:32:30 | | | |
| 2 | Catherine Yang | -165 word(s) | 1841 | 2022-05-07 08:44:05 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Song, J.; Liu, Y.; , .; Seidi Ghalehgah, F.; Xiao, H. Carbohydrate-Binding Modules of Potential Resources. Encyclopedia. Available online: https://encyclopedia.pub/entry/22680 (accessed on 04 March 2026).
Song J, Liu Y, , Seidi Ghalehgah F, Xiao H. Carbohydrate-Binding Modules of Potential Resources. Encyclopedia. Available at: https://encyclopedia.pub/entry/22680. Accessed March 04, 2026.
Song, Junlong, Yena Liu, , Farzad Seidi Ghalehgah, Huining Xiao. "Carbohydrate-Binding Modules of Potential Resources" Encyclopedia, https://encyclopedia.pub/entry/22680 (accessed March 04, 2026).
Song, J., Liu, Y., , ., Seidi Ghalehgah, F., & Xiao, H. (2022, May 07). Carbohydrate-Binding Modules of Potential Resources. In Encyclopedia. https://encyclopedia.pub/entry/22680
Song, Junlong, et al. "Carbohydrate-Binding Modules of Potential Resources." Encyclopedia. Web. 07 May, 2022.
Copy Citation
Carbohydrate-binding modules (CBMs) are a class of multi-module enzyme proteins and their function is to respond to bind to the carbohydrate substrate. Cellulose-binding domains (CBDs) are the earliest-discovered CBMs which were used to be catergozied based on their sequence homology. However, with the in-depth study of carbohydrate hydrolases, more modules in carbohydrate-active enzymes were discovered that could bind, in addition to cellulose, to other types of carbohydrates such as chitin, glucan, xylan, or starch.
carbohydrate-binding modules (CBM)
classification and configuration
CBM-substrate interactions
1. CBMs: Classification, Sources, Structures, and Functions
CBMs are widely distributed in nature [1] and are present in enzymes secreted by bacteria, fungi, and archaea [2]. Typical fungi sources are Trichoderma reesei [3], Caldanaerobius polysaccharolyticus [4], Rhizopus oryzae [5] and Polymyxa [6]. Fungi have developed to produce a set of glycoside hydrolases (GHs) and oxidoreductive enzymes, the synergistic action of which is required for enzymatic degradation of lignocellulose [7]. Bacteria commonly used in research are Clostridium thermocellum [8], maritima [9], Rhodothermus marinus [10], bacillus halodurans [11] and alcaligenes [12]. There are other microorganisms containing CBMs, such as actinomycetes [13]. Various types of CBMs are obtained from different microorganisms. Additionally, through genetic engineering, different expression vectors are constructed to obtain single or multiple CBMs, and used CBMs for substrate recognition and fiber treatment.
There are many ways to classify CBMs. Based on structural, functional similarities and the different ligand binding sites, CBMs can be divided into three types, namely, ‘surface-binding’ CBMs (type-A), ‘glycan-chain-binding’ CBMs (type-B), and ‘small-sugar-binding’ CBMs (type-C) [14]. For example, desired CBMs can be obtained by genetic engineering (the SUMO nobility tag can be added), as shown in Figure 1e.
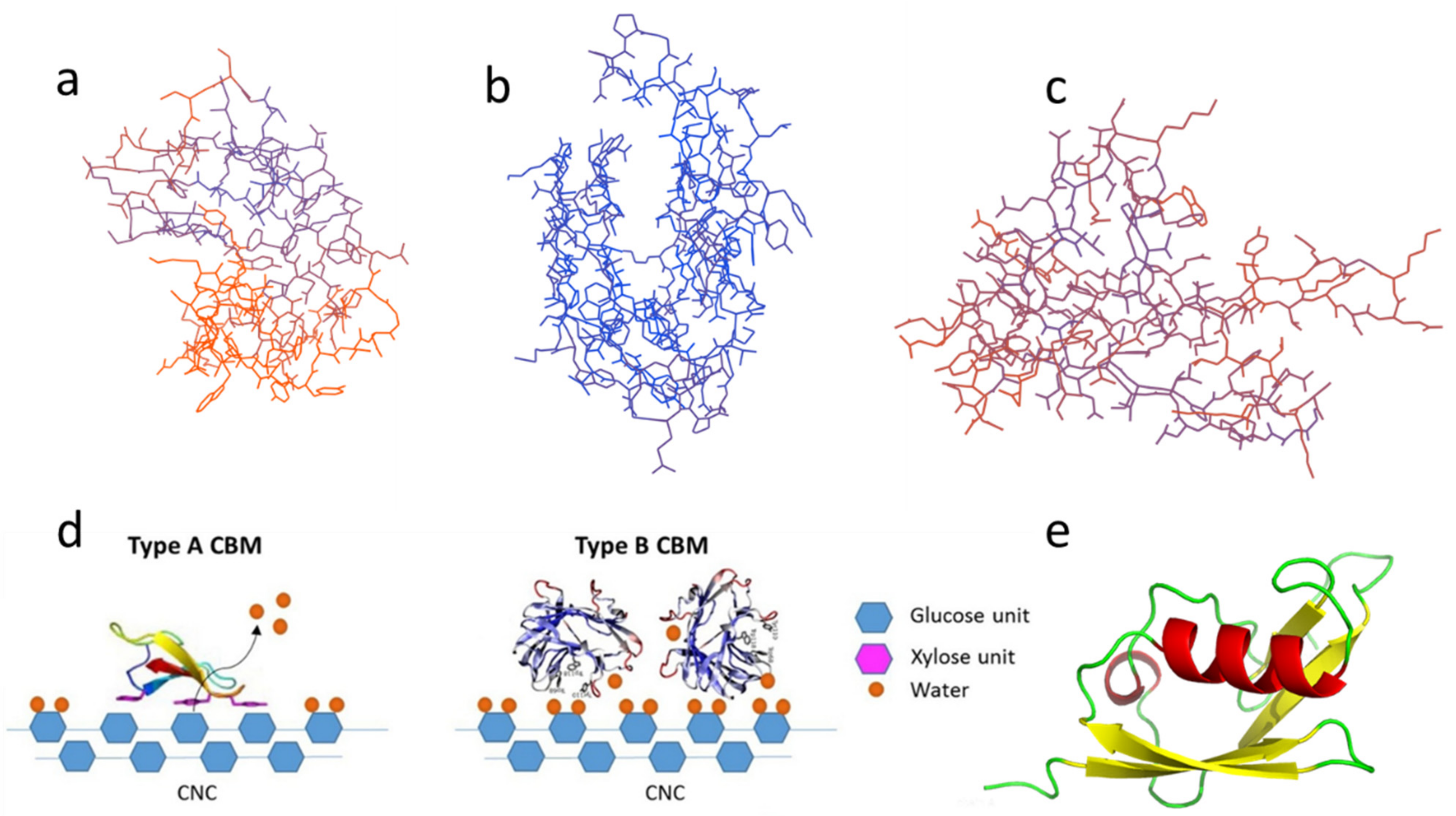
Figure 1. Different types of CBM: (a) Type-A, CBM3; (b) Type-B, CBM4; (c) Type-C, CBM9; (d) Schematic of binding of type-A (left) and type-B (right) CBMs on nanocrystalline cellulose (CNC) reprinted from Ref [15] with permission from Elsevier; (e) Type-A CBM1 with SUMO solubilizing label. The above structure diagrams are drawn using the base sequences from Table 1 through the Swiss model and Pymol.
1.1. Type-A CBMs
Type-A CBMs contain a hydrophobic surface, and the binding of CBMs tends to be distributed in a plane or near a plane, binding to the surface of crystal regions of carbohydrate substrate [16]. A schematic diagram of the binding of type-A CBMs on the fiber substrate is shown in Figure 1d. CBM1 and CBM3 are two typical type-A CBMs. Their 3D configurations are illustrated in Figure 1a [15]. CBM1, the smallest CBM currently found in nature, consists of approximately 36 residues and typically contains two or three disulfide bonds and a plane including three aligned aromatic residues along with several polar residues [17][18][19]. Uppsala University reported the first NMR spectrum of the CBM1 synthesized by solid peptide sequences from the most abundant cellulase in Trichoderma [20].
1.2. Type-B CBMs
The crystal structure of type-B CBMs shows that the protein of type-B CBMs often contains grooves or cracks of different depths, which is shown in Figure 1b [21]. They are grooved when the binding sites bind to amorphous cellulose [22] or mannan [23]. The schematic diagram of the binding of general type-B CBMs on the fiber substrate is displayed in Figure 1d [15]. Most type-B CBMs are produced by enzymes secreted by bacteria. The aromatic group only interacts with the free single-chain polysaccharide [8]. The crystal structure of CBM of cellobiohydrolase A derived from Clostridium thermocellum is the first discovered crystal structure of cellulase CBM4 [24]. And Alahuhta, et al. [25] have solved the X-ray structure of CelK CBM4 from C. thermocellum.
1.3. Type-C CBMs
The typical configurations of type-C CBMs, including CBM9, 14, etc., are illustrated in Figure 1c. Type-C CBMs mainly interact with the end of the polysaccharide chain. Due to steric hindrance, only monosaccharides, disaccharides, trisaccharides, or the terminal sugar group of polysaccharides bind to type-C CBMs [26]. Type-C CBMs was first known from lectins, which are widely found in animals, plants, and microorganisms, and can bind to free sugars in solution. A lectin contains multiple CBMs and can selectively bind to a specific glycosyl [27]. At present, there are few related studies on type-C CBMs.
1.4. Other Classification Methods
Other classification methods can be based on the family and folding configuration. In terms of configuration, members of the large majority of CBM families are β-conformations, including β-sandwich, β-Strefoil, Cysteine knot, Unique, OD fold, and Hevein fold [28]. What is interesting is that different types of CBMs can coexist in a single protein, which suggests that current classifications may not cover all functional classifications of CBMs found in nature [29]. And more and more CBMs from different sources are being discovered. The structures, functions, and characteristics of CBMs lay a foundation for CBMs to conjugate or fuse with other polymers and eventually apply in substrate recognition and fiber treatment.
2. Fiber Treatment Using CBMs
In recent years, with the boycott of plastic products, the demand for fiber materials has increased. However, due to the insufficient strength (especially wet strength) of packaging, paper straws, and the requirements for cleaner production, new biological treatment have gradually attracted the attention of researchers. Among them, application of single or multiple CBMs in fiber processing has extensively been utilized for improving the fiber properties. Treating cellulose fibers with CBMs can change their interfacial properties [30]. CBMs were fused to engineering enzymes/proteins for improved biological activity; or either used alone or conjugated with other reagents for enhanced wood and fiber treatment performance. Using CBM-based polymers to treat fibers to gain improvement of mechanical properties of fiber (secondary fiber) is an emerging area that should pay much attention [31].
2.1. Use CBMs Alone in Fiber Treatment
Pala [32] first used separate CBM in papermaking to improve the water filtration and mechanical strength of secondary fiber paper. It showed that CBMs obtained by proteolysis of T. reesei cellulase can alter the drainage capacity of recycled pulp [33]. Shoseyov, et al. [34] and Laaksonen, et al. [35] developed biofunctional CBMs by genetic engineering and obtained paper-based materials with high mechanical strength. The adhesion domain was constructed by CBMs and amphiphilic hydrophobic protein (HFBI). A hydrophobic AFM tip can contact and lift a single fusion protein from the functionalized HFBI terminal through hydrophobic interactions between the tip surface and the HFBI hydrophobic patch [35]. Shi, et al. [36] constructed four recombinant CBMs, CBM3-GS(polypeptide (G4S)3)-CBM1, CBM3-NL(native linker from CBH1-1)-CBM1, CBM3-GS-CBM3, and CBM1-NL-CBM1, the mechanical properties of paper were all enhanced. The folding resistance and tensile strength of paper increased by 27.4% and 15.5% after adding CBM3-GS-CBM3, and after the addition of CBM1-NL-CBM1, the paper tensile strength, elongation, and folding resistance was increased by 12.6%, 8.8%, and 16.7%, respectively. Among them, the improvement of tensile strength and folding resistance facilitate the use of containerboard paper. The fiber agglomerations disappeared after CBMs treatment [33]. CBMs destroyed the aggregates dispersed on the larger fiber surface during drying. This is an interfacial phenomenon. CBMs treatment may reduce fiber interaction (fiber separation observed by SEM) through spatial and hydrophobic effects. Therefore, in the wet state, CBMs may have a better effect on fibers. However, the use of CBMs alone is expensive and cannot fufill are the desire requirements. Therefore, the researchers explored of the comination of other treaments along with CBMs to improve the fibers’ properties. Pretreated the fibers with CBMs and refining, then used water retention value (WRV), SEM, and aspect ratio to observe the change of the fiber. The results showed that using CBMs to more accurately conjecture enzyme accessibility, and it was found that refining did not significantly improve enzyme accessibility at the microfiber level of the cellulose substrate. Later, researchers began to study the conjugated additives, to achieve both performance and economic satisfaction.
2.2. CBMs Conjugated with Other Polymers for Fiber Treatment
CBMs can conjugate with other proteins or polymers because of their flexibility and specificity of CBMs. Protein side-chains contain many groups, such as amino, carboxyl, and hydroxyl groups [39]. Complex can be produced by common methods of blending (electrostatic attraction), and conjugation [40]. Many researchers began to construct conjugated systems of CBMs and polymers. CBM can be conjugated with various compounds such as polyethylene glycol (PEG), and anionic polyacrylamide (APAM) [41]. Machado [37] studied the adsorption of a CBM3 from the Clostridium thermocellum scaffolding protein (Cip A) to cellulose. The Carbohydrate binding domain-polyethylene glycol (CBM-PEG) module was constructed and the effect of this structure on the paper properties was studied (see Figure 3c). CBM-PEG improved the drainage capacity, but does not affect the mechanical properties of the paper which is due to the high water-binding capacity of PEG [42]. CBM-PEG improved the drainability of E. globulus and P. sylvestris pulps without affecting the physical properties of the paper [43]. Kitaoka and Tanaka [41] conjugated the CBM with APAM to improve the fiber binding, the results showed that both the dry tensile index and the wet tensile index were improved. However, both the fiber and the APAM are negatively charged, and the APAM is mostly used as a dispersant in the paper industry, in this case, there is still an improvement in mechanical properties, which can show the superiority of CBM for fiber binding.
The advantages of using independent CBM in fiber processing include the diversity of CBMs and avoiding the strength loss of using whole enzymes due to the catalytic activity of CD. More importantly, the fusion method with other polymers significantly reduces the amount of CBMs required and therefore reduces the costs. However, mass and economical production, preservation, and transportation of CBMs are still critical prerequisites for CBMs’ industrial applications. The current related work is very important because of the increased demand and performance requirements for paper products [44]. Further progress in this area is required to provide more environmentally friendly and more economical additives to improve fiber strength. Meanwhile, there are a few studies on the use of CBMs for nanocellulose materials, such as bacterial cellulose and microcrystalline cellulose materials [45]. This is also a major research direction because the structural properties of CBMs have the potential to alter the brittleness of nanocellulose materials [46]. Nanocellulose materials can be used in Pickering emulsions [47], ultrafiltration membrane [48][49] and paper straws [50].
2.3. Other Functions
In addition to the above effects on cellulose, the fusion of CBMs with other enzymes can also change biochemical characteristics and improve catalytic performance. And the CBMs of some thermophilic bacteria have high stability and belong to the thermostable domain. Studies have shown that fusion of thermostability domains to unstable protein domains can improve the thermostability of the latter [51][52]. Chhabra and Kelly [53] first reported the hyperthermophilic CBM fused to hyperthermophilic endoglucanase. The fusion protein was active on crystalline cellulose and the activity against microcrystalline cellulose was higher than that of the parent endoglucanase at 80 °C. Kavoosi, et al. [54] evaluated the impact of linker design on fusion protein production and performance. Liu, et al. [55] constructed an artificial bifunctional enzyme containing carbonic anhydrase(CA) from Neisseria gonorrhoeae and the CBM from Clostridium thermocellum with His6 tag, which can capture carbon dioxide from flue gas. As for the improvement of catalytic efficiency, Kittur, et al. [56] increased the catalytic activity of xylanase from Thermotoga Maritima for soluble xylan by fusion of CBM2. For optimizing the catalytic activity of Cyclodextrin glycosyltransferase (CGTase). It is an important industrial enzyme for the production of cyclodextrins (CDs) from starch by intramolecular transglycosylation. CGTase of Geobacillus sp. was fused with the CBM20 of the Bacillus circulans strain 251 CGTase [57]. There seemed to be much room for improving its enzymological properties, such as improving its catalytic efficiency and substrate affinity, by replacing the domain of wild-type structural domain with a suitable CBM [58].
References
- Varghese, B.; McKee, G.; Alexandrov, V. Automating fault tolerance in high-performance computational biological jobs using multi-agent approaches. Comput. Biolog. Medicine 2014, 48, 28–41.
- Minami, A.; Ozaki, T.; Liu, C.; Oikawa, H. Cyclopentane-forming di/sesterterpene synthases: Widely distributed enzymes in bacteria, fungi, and plants. Nat. Prod. Rep. 2018, 35, 1330–1346.
- Stojakowska, A.; Michalska, K.; Malarz, J. Simultaneous quantification of eudesmanolides and thymol derivatives from tissues of Inula helenium and I-royleana by reversed-phase high-performance liquid chromatography. Phytochem. Analys. 2006, 17, 157–161.
- Bae, B.; Ohene-Adjei, S.; Kocherginskaya, S.; Mackie, R.I.; Spies, M.A.; Cann, I.K.O.; Nair, S.K. Molecular basis for the selectivity and specificity of ligand recognition by the family 16 carbohydrate-binding modules from Thermoanaerobacterium polysaccharolyticum ManA. J. Biolog. Chem. 2008, 283, 12415–12425.
- Liu, Y.-N.; Lai, Y.-T.; Chou, W.-I.; Chang, M.D.-T.; Lyu, P.-C. Solution structure of family 21 carbohydrate-binding module from Rhizopus oryzae glucoamylase. Biochem. J. 2007, 403, 21–30.
- Jamal-Talabani, S.; Boraston, A.B.; Turkenburg, J.P.; Tarbouriech, N.; Ducros, V.M.A.; Davies, G.J. Ab initio structure determination and functional characterization of CBM36: A new family of calcium-dependent carbohydrate binding modules. Structure 2004, 12, 1177–1187.
- Varnai, A.; Makela, M.R.; Djajadi, D.T.; Rahikainen, J.; Hatakka, A.; Viikari, L. Carbohydrate-binding modules of fungal cellulases: Occurrence in nature, function, and relevance in industrial biomass conversion. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 88, pp. 103–165.
- Kljun, A.; Benians, T.A.S.; Goubet, F.; Meulewaeter, F.; Knox, J.P.; Blackburn, R.S. Comparative Analysis of Crystallinity Changes in Cellulose I Polymers Using ATR-FTIR, X-ray Diffraction, and Carbohydrate-Binding Module Probes. Biomacromolecules 2011, 12, 4121–4126.
- Boraston, A.B.; Revett, T.J.; Boraston, C.M.; Nurizzo, D.; Davies, G.J. Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate binding module, TmCBM27. Structure 2003, 11, 665–675.
- Simpson, P.J.; Jamieson, S.J.; Abou-Hachem, M.; Karlsson, E.N.; Gilbert, H.J.; Holst, O.; Williamson, M.P. The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase. Biochemistry 2002, 41, 5712–5719.
- Boraston, A.B.; Healey, M.; Klassen, J.; Ficko-Blean, E.; van Bueren, A.L.; Law, V. A structural and functional analysis of alpha-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition. J. Biolog. Chem. 2006, 281, 587–598.
- Hashimoto, H.; Tamai, Y.; Okazaki, F.; Tamaru, Y.; Shimizu, T.; Araki, T.; Sato, M. The first crystal structure of a family 31 carbohydrate-binding module with affinity to beta-1, 3-xylan. FEBS Lett. 2005, 579, 4324–4328.
- Suzuki, N.; Fujimoto, Z.; Kim, Y.-M.; Momma, M.; Kishine, N.; Suzuki, R.; Suzuki, S.; Kitamura, S.; Kobayashi, M.; Kimura, A.; et al. Structural Elucidation of the Cyclization Mechanism of alpha-1,6-Glucan by Bacillus circulans T-3040 Cycloisomaltooligosaccharide Glucanotransferase. J. Biolo. Chem. 2014, 289, 12040–12051.
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochemical J. 2004, 382, 769–781.
- Liu, T.; Zhang, Y.; Lu, X.; Wang, P.; Zhang, X.; Tian, J.; Wang, Q.; Song, J.; Jin, Y.; Xiao, H. Binding affinity of family 4 carbohydrate binding module on cellulose films of nanocrystals and nanofibrils. Carbohydr. Polym. 2021, 251, 116725.
- Linder, M.; Lindeberg, G.; Reinikainen, T.; Teeri, T.T.; Pettersson, G.R. The difference in affinity between two fungal cellulose-binding domains is dominated by a single amino acid substitution. FEBS Lett. 1995, 372, 96–98.
- Carvalho, C.C.; Phan, N.N.; Chen, Y.; Reilly, P.J. Carbohydrate-binding module tribes. Biopolymers 2015, 103, 203–214.
- Zhang, Y.; Wang, X.; Wang, P.; Song, J.; Jin, Y.; Rojas, O.J. Interactions between type A carbohydrate binding modules and cellulose studied with a quartz crystal microbalance with dissipation monitoring. Cellulose 2020, 27, 3661–3675.
- Zhang, Y.; Yang, F.; Hu, F.; Song, J.; Wu, S.; Jin, Y. Binding preference of family 1 carbohydrate binding module on nanocrystalline cellulose and nanofibrillar cellulose films assessed by quartz crystal microbalance. Cellulose 2018, 25, 3327–3337.
- Kraulis, J.; Clore, G.M.; Nilges, M.; Jones, T.A.; Pettersson, G.; Knowles, J.; Gronenborn, A.M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry 1989, 28, 7241–7257.
- Boraston, A.B.; Nurizzo, D.; Notenboom, V.; Ducros, V.; Rose, D.R.; Kilburn, D.G.; Davies, G.J. Differential Oligosaccharide Recognition by Evolutionarily-related β-1,4 and β-1,3 Glucan-binding Modules. J. Molecul. Biol. 2002, 319, 1143–1156.
- Zhu, J.; Zhao, Y.; Hu, Q.; Zhang, Y.; Shao, T.; Fan, B.; Jiang, Y.; Chen, Z.; Zhao, M. Coalbed Methane Production Model Based on Random Forests Optimized by a Genetic Algorithm. ACS Omega 2022, 7, 13083–13094.
- Jamal, S.; Nurizzo, D.; Boraston, A.B.; Davies, G.J. X-ray crystal structure of a non-crystalline cellulose-specific carbohydrate-binding module: CBM28. J. Mol. Biol. 2004, 339, 253–258.
- Alahuhta, M.; Xu, Q.; Bomble, Y.J.; Brunecky, R.; Adney, W.S.; Ding, S.-Y.; Himmel, M.E.; Lunin, V.V. The Unique Binding Mode of Cellulosomal CBM4 from Clostridium thermocellum Cellobiohydrolase A. J. Mol. Biol. 2010, 402, 374–387.
- Alahuhta, M.; Luo, Y.; Ding, S.Y.; Himmel, M.E.; Lunin, V.V. Structure of CBM4 from Clostridium thermocellum cellulase K. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 527–530.
- Notenboom, V.; Boraston, A.B.; Kilburn, D.G.; Rose, D.R. Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima Xylanase 10A in native and ligand-bound forms. Biochemistry 2001, 40, 6248–6256.
- Sharon, N.; Lis, H. Lectins as cell recognition molecules. Science 1989, 246, 227–234.
- Freeman, H.; Srinivasan, S.; Das, D.; Stayton, P.S.; Convertine, A.J. Fully synthetic macromolecular prodrug chemotherapeutics with EGFR targeting and controlled camptothecin release kinetics. Polym. Chem. 2018, 9, 5224–5233.
- Thiers, F.A.; Burgess, B.J.; Nadol, J.B., Jr. Reciprocal innervation of outer hair cells in a human infant. J. Assoc. Res. Otolaryngol. 2002, 3, 269–278.
- Shoseyov, O.; Shani, Z.; Levy, I. Carbohydrate binding modules: Biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 2006, 70, 283–295.
- Saw, S.K.; Akhtar, K.; Yadav, N.; Singh, A.K. Hybrid Composites Made from Jute/Coir Fibers: Water Absorption, Thickness Swelling, Density, Morphology, and Mechanical Properties. J. Nat. Fibers 2014, 11, 39–53.
- Pala, H. Enzymatic upgrade of old paperboard containers. Enzym. Microb. Technol. 2001, 29, 2274–2279.
- Pinto, R.; Moreira, S.; Mota, M.; Gama, M. Studies on the cellulose-binding domains adsorption to cellulose. Langmuir 2004, 20, 1409–1413.
- Shoseyov, O.; Levy, I.; Shani, Z.; Mansfield, S.D. Modulation of wood fibers and paper by cellulose-binding domains. In Applications of Enzymes to Lignocellulosics; Mansfield, S.D., Saddler, J.N., Eds.; ACS Publications: Washington, DC, USA, 2003; Volume 855, pp. 116–131.
- Laaksonen, P.; Walther, A.; Malho, J.-M.; Kainlauri, M.; Ikkala, O.; Linder, M.B. Genetic Engineering of Biomimetic Nanocomposites: Diblock Proteins, Graphene, and Nanofibrillated Cellulose. Angew. Chem. Int. Ed. 2011, 50, 8688–8691.
- Shi, X.; Zheng, F.; Pan, R.; Wang, J.; Ding, S. Engineering and Comparative Characteristics of Double Carbohydrate Binding Modules as a Strength Additive for Papermaking Applications. Bioresources 2014, 9, 3117–3131.
- Machado, J.; Araújo, A.; Pinto, R.; Gama, F.M. Studies on the interaction of the carbohydrate binding module 3 from the Clostridium thermocellum CipA scaffolding protein with cellulose and paper fibres. Cellulose 2009, 16, 817–824.
- Aissa, K.; Karaaslan, M.A.; Renneckar, S.; Saddler, J.N. Functionalizing Cellulose Nanocrystals with Click Modifiable Carbohydrate-Binding Modules. Biomacromolecules 2019, 20, 3087–3093.
- Clementi, C.; Garcı́a, A.E.; Onuchic, J.N. Interplay Among Tertiary Contacts, Secondary Structure Formation and Side-chain Packing in the Protein Folding Mechanism: All-atom Representation Study of Protein L. J. Mol. Biol. 2003, 326, 933–954.
- Sam, S.; Touahir, L.; Salvador Andresa, J.; Allongue, P.; Chazalviel, J.N.; Gouget-Laemmel, A.C.; Henry de Villeneuve, C.; Moraillon, A.; Ozanam, F.; Gabouze, N.; et al. Semiquantitative study of the EDC/NHS activation of acid terminal groups at modified porous silicon surfaces. Langmuir 2010, 26, 809–814.
- Kitaoka, T.; Tanaka, H. Novel paper strength additive containing cellulose-binding domain of cellulase. J. Wood Sci. 2001, 47, 322–324.
- Vincent, F.; Dal Molin, D.; Weiner, R.M.; Bourne, Y.; Henrissat, B. Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown. FEBS Lett. 2010, 584, 1577–1584.
- Aissa, K.; Novy, V.; Nielsen, F.; Saddler, J. Use of carbohydrate binding modules to elucidate the relationship between fibrillation, hydrolyzability, and accessibility of cellulosic substrates. ACS Sustain. Chem. Eng. 2019, 7, 1113–1119.
- Barbosa, M.; Simoes, H.; Pinto, S.N.; Macedo, A.S.; Fonte, P.; Prazeres, D.M.F. Fusions of a carbohydrate binding module with the small cationic hexapeptide RWRWRW confer antimicrobial properties to cellulose-based materials. Acta Biomater. 2022, 143, 216–232.
- Koskela, S.; Wang, S.; Xu, D.; Yang, X.; Li, K.; Berglund, L.A.; McKee, L.S.; Bulone, V.; Zhou, Q. Lytic polysaccharide monooxygenase (LPMO) mediated production of ultra-fine cellulose nanofibres from delignified softwood fibres. Green Chem. 2019, 21, 5924–5933.
- Griffo, A.; Rooijakkers, B.J.M.; Haehl, H.; Jacobs, K.; Linder, M.B.; Laaksonen, P. Binding Forces of Cellulose Binding Modules on Cellulosic Nanomaterials. Biomacromolecules 2019, 20, 769–777.
- Li, Q.; Ma, Z.; Meng, D.; Sui, X.; You, C. Facile biosynthesis of synthetic crystalline cellulose nanoribbon from maltodextrin through a minimized two-enzyme phosphorylase cascade and its application in emulsion. J. Biotechnol. 2021, 332, 54–60.
- Mautner, A.; Bismarck, A. Bacterial nanocellulose papers with high porosity for optimized permeance and rejection of nm-sized pollutants. Carbohydr. Polym. 2021, 251, 117130.
- Mautner, A.; Lee, K.-Y.; Tammelin, T.; Mathew, A.P.; Nedoma, A.J.; Li, K.; Bismarck, A. Cellulose nanopapers as tight aqueous ultra-filtration membranes. React. Funct. Polym. 2015, 86, 209–214.
- Wang, S.; Jiang, F.; Xu, X.; Kuang, Y.; Fu, K.; Hitz, E.; Hu, L. Super-Strong, Super-Stiff Macrofibers with Aligned, Long Bacterial Cellulose Nanofibers. Adv. Mater. 2017, 29, 1702498.
- Pierre, B.; Labonte, J.W.; Xiong, T.; Aoraha, E.; Williams, A.; Shah, V.; Chau, E.; Helal, K.Y.; Gray, J.J.; Kim, J.R. Molecular Determinants for Protein Stabilization by Insertional Fusion to a Thermophilic Host Protein. Chembiochem 2015, 16, 2392–2402.
- Ribeiro, L.F.; Furtado, G.P.; Lourenzoni, M.R.; Costa-Filho, A.J.; Santos, C.R.; Peixoto Nogueira, S.C.; Betini, J.A.; Polizeli, M.d.L.T.M.; Murakami, M.T.; Ward, R.J. Engineering Bifunctional Laccase-Xylanase Chimeras for Improved Catalytic Performance. J. Biol. Chem. 2011, 286, 43026–43038.
- Chhabra, S.R.; Kelly, R.M. Biochemical characterization of Thermotoga maritima endoglucanase Ce174 with and without a carbohydrate binding module (CBM). FEBS Lett. 2002, 531, 375–380.
- Kavoosi, M.; Creagh, A.L.; Kilburn, D.G.; Haynes, C.A. Strategy for selecting and characterizing linker peptides for CBM9-tagged fusion proteins expressed in Escherichia coli. Biotechnol. Bioeng. 2007, 98, 599–610.
- Liu, Z.; Bartlow, P.; Dilmore, R.M.; Soong, Y.; Pan, Z.; Koepsel, R.; Ataai, M. Production, Purification, and Characterization of a Fusion Protein of Carbonic Anhydrase from Neisseria gonorrhoeae and Cellulose Binding Domain from Clostridium thermocellum. Biotechnol. Prog. 2009, 25, 68–74.
- Kittur, F.S.; Mangala, S.L.; Rus’d, A.A.; Kitaoka, M.; Tsujibo, H.; Hayashi, K. Fusion of family 2b carbohydrate-binding module increases the catalytic activity of a xylanase fromThermotoga maritimato soluble xylan. FEBS Lett. 2003, 549, 147–151.
- Jia, X.; Guo, Y.; Lin, X.; You, M.; Lin, C.; Chen, L.; Chen, J. Fusion of a family 20 carbohydrate-binding module (CBM20) with cyclodextrin glycosyltransferase of Geobacillus sp CHB1 improves catalytic efficiency. J. Basic Microbiol. 2017, 57, 471–480.
- Ara, K.Z.G.; Lundemo, P.; Fridjonsson, O.H.; Hreggvidsson, G.O.; Adlercreutz, P.; Karlsson, E.N. A CGTase with high coupling activity using γ-cyclodextrin isolated from a novel strain clustering under the genus Carboxydocella. Glycobiology 2015, 25, 514–523.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.9K
Entry Collection:
Tight Junction and Its Proteins
Revisions:
2 times
(View History)
Update Date:
07 May 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





