
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mikhail Soloviev | -- | 3796 | 2022-05-04 21:31:37 | | | |
| 2 | Vivi Li | + 7 word(s) | 3803 | 2022-05-05 05:22:16 | | |
Video Upload Options
Globally, colorectal cancer (CRC) is the third most common cancer, with 1.4 million new cases and over 700,000 deaths per annum. Despite being one of the most common cancers, few molecular approaches to detect CRC exist. Carcinoembryonic antigen (CEA) is a known serum biomarker that is used in CRC for monitoring disease recurrence or response to treatment. However, it can also be raised in multiple benign conditions, thus having no value in early detection or screening for CRC. Molecular biomarkers play an ever-increasing role in the diagnosis, prognosis, and outcome prediction of disease, however, only a limited number of biomarkers are available and none are suitable for early detection and screening of CRC. A PCR-based Epi proColon® blood plasma test for the detection of methylated SEPT9 has been approved by the USFDA for CRC screening in the USA, alongside a stool test for methylated DNA from CRC cells. However, these are reserved for patients who decline traditional screening methods. There remains an urgent need for the development of non-invasive molecular biomarkers that are highly specific and sensitive to CRC and that can be used routinely for early detection and screening. A molecular approach to the discovery of CRC biomarkers focuses on the analysis of the transcriptome of cancer cells to identify differentially expressed genes and proteins. A systematic search of the literature yielded over 100 differentially expressed CRC molecular markers, of which the vast majority are overexpressed in CRC. In terms of function, they largely belong to biological pathways involved in cell division, regulation of gene expression, or cell proliferation, to name a few.
1. Introduction
2. Obstacles and Limitations to the Use of Biomarkers as a Screening Tool
3. Current Advances in CRC Biomarker Detection
3.1. DNA-Based Molecular Markers and Tests
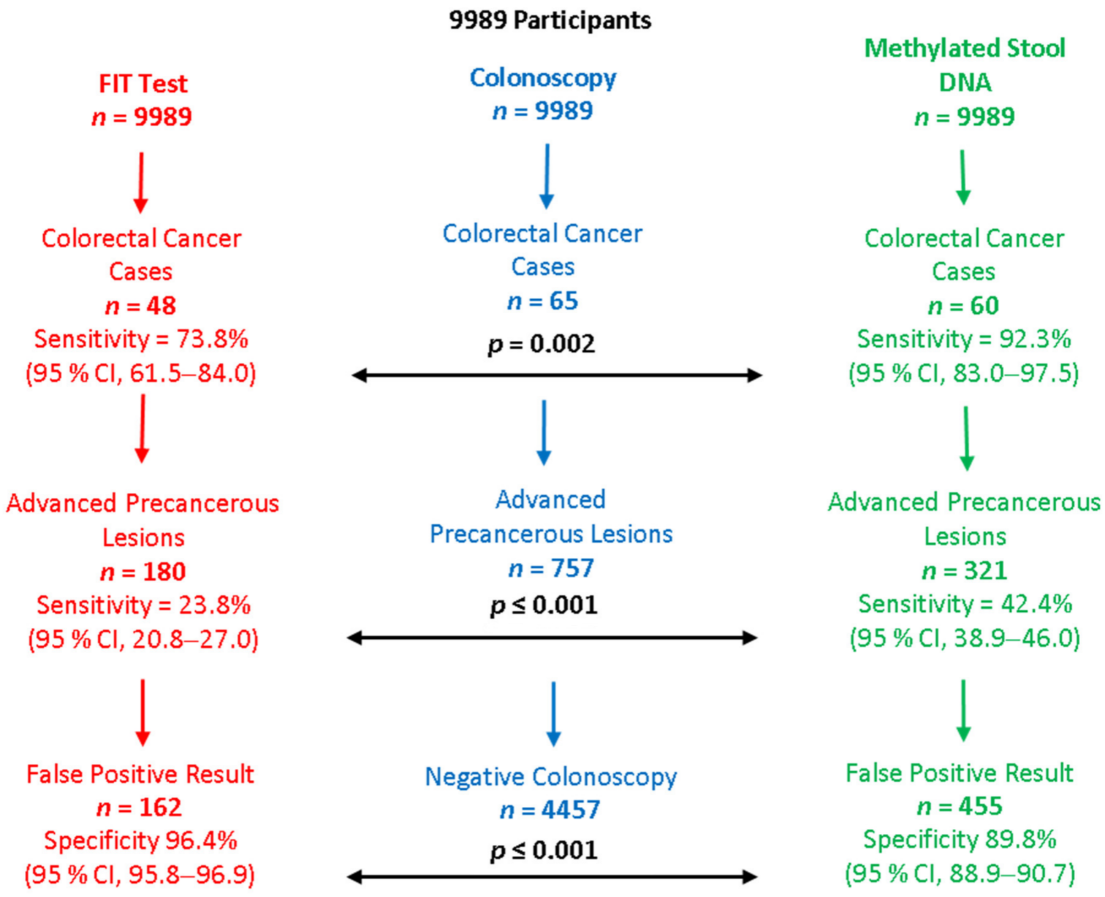
3.2. Circulating Tumor Cells
3.3. microRNAs and Other Non-Coding RNAs
3.4. Differential Gene and Protein Expression in CRC
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386.
- Abd Algfoor, Z.; Shahrizal Sunar, M.; Abdullah, A.; Kolivand, H. Identification of metabolic pathways using pathfinding approaches: A systematic review. Brief. Funct. Genom. 2017, 16, 87–98.
- Atlasi, Y.; Noori, R.; Marolin, I.; Franken, P.; Brandao, J.; Biermann, K.; Collini, P.; Grigorian, M.; Lukanidin, E.; Ambartsumian, N.; et al. The role of S100a4 (Mts1) in Apc- and Smad4-driven tumour onset and progression. Eur. J. Cancer 2016, 68, 114–124.
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416.
- Li, Y.; Yuan, Y. Alternative RNA splicing and gastric cancer. Mutat. Res. 2017, 773, 263–273.
- Muller, M.F.; Ibrahim, A.E.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. Int. J. Pathol. 2016, 469, 125–134.
- Nicholson, B.D.; James, T.; East, J.E.; Grimshaw, D.; Paddon, M.; Justice, S.; Oke, J.L.; Shine, B. Experience of adopting faecal immunochemical testing to meet the NICE colorectal cancer referral criteria for low-risk symptomatic primary care patients in Oxfordshire, UK. Frontline Gastroenterol. 2019, 10, 347–355.
- O’Connor, L.; Gilmour, J.; Bonifer, C. The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-specific Transcriptional Regulation and in Disease. Yale J. Biol. Med. 2016, 89, 513–525.
- Cancer Research, UK. Bowel Cancer Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer (accessed on 12 October 2021).
- Huang, Q.; Li, S.; Cheng, P.; Deng, M.; He, X.; Wang, Z.; Yang, C.-H.; Zhao, X.-Y.; Huang, J. High expression of anti-apoptotic protein Bcl-2 is a good prognostic factor in colorectal cancer: Result of a meta-analysis. World J. Gastroenterol. 2017, 23, 5018–5033.
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691.
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917.
- Whiffin, N.; Hosking, F.J.; Farrington, S.M.; Palles, C.; Dobbins, S.E.; Zgaga, L.; Lloyd, A.; Kinnersley, B.; Gorman, M.; Tenesa, A.; et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum. Mol. Genet. 2014, 23, 4729–4737.
- Nandakumar, G.; Morgan, J.A.; Silverberg, D.; Steinhagen, R.M. Familial polyposis coli: Clinical manifestations, evaluation, management and treatment. Mt. Sinai. J. Med. 2004, 71, 384–391.
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531.
- Shussman, N.; Wexner, S.D. Colorectal polyps and polyposis syndromes. Gastroenterol. Rep. 2014, 2, 1–15.
- Ahnen, D.J. The American College of Gastroenterology Emily Couric Lecture—The adenoma-carcinoma sequence revisited: Has the era of genetic tailoring finally arrived? Am. J. Gastroenterol. 2011, 106, 190–198.
- Wieszczy, P.; Kaminski, M.; Franczyk, R.; Loberg, M.; Kobiela, J.; Rupinska, M.; Kocot, B.; Rupinski, M.; Holme, O.; Wojciechowska, U.; et al. Colorectal Cancer Incidence and Mortality After Removal of Adenomas During Screening Colonoscopies. Gastroenterology 2020, 158, 875–883.e5.
- Au, F.C.; Stein, B.; Tang, C.K. Carcinoembryonic antigen levels in colonic lesions. Am. J. Surg. 1986, 151, 61–64.
- Scarà, S.; Bottoni, P.; Scatena, R. CA 19-9: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 247–260.
- Pavai, S.; Yap, S.F. The clinical significance of elevated levels of serum CA 19-9. Med. J. Malays. 2003, 58, 667–672.
- Tsen, A.; Barbara, M.; Rosenkranz, L. Dilemma of elevated CA 19-9 in biliary pathology. Pancreatology 2018, 18, 862–867.
- Bottoni, P.; Scatena, R. The Role of CA 125 as Tumor Marker: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 229–244.
- Fiala, L.; Bob, P.; Raboch, J. Oncological markers CA-125, CA 19-9 and endometriosis. Medicine 2018, 97, e13759.
- Presti, J., Jr.; Alexeeff, S.; Horton, B.; Prausnitz, S.; Avins, A.L. Changes in Prostate Cancer Presentation Following the 2012 USPSTF Screening Statement: Observational Study in a Multispecialty Group Practice. J. Gen. Intern. Med. 2019, 35, 1368–1374.
- Kuppusamy, S.; Gillatt, D. Managing patients with acute urinary retention. Practitioner 2011, 255, 21–24.
- CDC. Colorectal Cancer Screening Tests. Available online: https://www.cdc.gov/cancer/colorectal/basic_info/screening/tests.htm (accessed on 14 October 2021).
- American Cancer Society. Colorectal Cancer Staging. Available online: https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/acs-recommendations.html (accessed on 24 October 2021).
- Heisser, T.; Weigl, K.; Hoffmeister, M.; Brenner, H. Age-specific sequence of colorectal cancer screening options in Germany: A model-based critical evaluation. PLoS Med. 2020, 17, e1003194.
- Pellat, A.; Deyra, J.; Coriat, R.; Chaussade, S. Results of the national organised colorectal cancer screening program with FIT in Paris. Sci. Rep. 2018, 8, 4162.
- Danish Cancer Society. Colon Cancer Screening Guidelines. Available online: https://www.cancer.dk/international/english/screening-colon-cancer-english (accessed on 21 October 2021).
- NHS. The NICE FIT Study. Available online: https://www.nicefitstudy.com/ (accessed on 10 October 2021).
- Cunningham, C.; Leong, K.; Clark, S.; Plumb, A.; Taylor, S.; Geh, I.; Karandikar, S.; Moran, B. Association of Coloproctology of Great Britain & Ireland (ACPGBI): Guidelines for the Management of Cancer of the Colon, Rectum and Anus (2017)—Diagnosis, Investigations and Screening. Colorectal Dis. Off. J. Assoc. Coloproctol. Great Br. Irel. 2017, 19 (Suppl. S1), 9–17.
- Imperiale, T.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014, 370, 1287–1297.
- Vakil, N.; Ciezki, K.; Huq, N.; Singh, M. Multitarget stool DNA testing for the prevention of colon cancer: Outcomes in a large integrated healthcare system. Gastrointest. Endosc. 2020, 92, 334–341.
- Cheng, Y.C.; Wu, P.H.; Chen, Y.J.; Yang, C.H.; Huang, J.L.; Chou, Y.C.; Chang, P.K.; Wen, C.C.; Jao, S.W.; Huang, H.H.; et al. Using Comorbidity Pattern Analysis to Detect Reliable Methylated Genes in Colorectal Cancer Verified by Stool DNA Test. Genes 2021, 12, 1539.
- Liu, C.; Xu, L.; Li, W.; Jie, M.; Xue, W.; Yu, W. Multiple Biomarker-Combined Screening for Colorectal Cancer Based on Bisulfate Conversion-Free Detection of Fecal DNA Methylation. BioMed Res. Int. 2021, 2021, 1479748.
- Exact Sciences. Cologuard Was Superior to a Leading FIT* in Detecting Colorectal Cancer (CRC) and Precancer. Available online: https://www.cologuardhcp.com/about/cologuard-vs-fit (accessed on 3 February 2022).
- Shirley, M. Epi proColon® for Colorectal Cancer Screening: A Profile of Its Use in the USA. Mol. Diagn. Ther. 2020, 24, 497–503.
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 2014, 60, 1183–1191.
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133.
- U.S. FDA. Epi proColon® Screening Approval. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf13/P130001C.pdf (accessed on 24 October 2021).
- Amir, S.; Golan, M.; Mabjeesh, N.J. Targeted knockdown of SEPT9_v1 inhibits tumor growth and angiogenesis of human prostate cancer cells concomitant with disruption of hypoxia-inducible factor-1 pathway. Mol. Cancer Res. 2010, 8, 643–652.
- Kim, D.S.; Hubbard, S.L.; Peraud, A.; Salhia, B.; Sakai, K.; Rutka, J.T. Analysis of mammalian septin expression in human malignant brain tumors. Neoplasia 2004, 6, 168–178.
- Burrows, J.F.; Chanduloy, S.; McIlhatton, M.A.; Nagar, H.; Yeates, K.; Donaghy, P.; Price, J.; Godwin, A.K.; Johnston, P.G.; Russell, S.H. Altered expression of the septin gene, SEPT9, in ovarian neoplasia. J. Pathol. 2003, 201, 581–588.
- Montagna, C.; Lyu, M.-S.; Hunter, K.; Lukes, L.; Lowther, W.; Reppert, T.; Hissong, B.; Weaver, Z.; Ried, T. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003, 63, 2179–2187.
- Kojima, K.; Sakai, I.; Hasegawa, A.; Niiya, H.; Azuma, T.; Matsuo, Y.; Fujii, N.; Tanimoto, M.; Fujita, S. FLJ10849, a septin family gene, fuses MLL in a novel leukemia cell line CNLBC1 derived from chronic neutrophilic leukemia in transformation with t(4;11)(q21;q23). Leukemia 2004, 18, 998–1005.
- Oh, T.; Kim, N.; Moon, Y.; Kim, M.S.; Hoehn, B.D.; Park, C.H.; Kim, T.S.; Kim, N.K.; Chung, H.C.; An, S. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J. Mol. Diagn. 2013, 15, 498–507.
- Zhang, L.; Dong, L.; Lu, C.; Huang, W.; Yang, C.; Wang, Q.; Wang, Q.; Lei, R.; Sun, R.; Wan, K.; et al. Methylation of SDC2/TFPI2 and Its Diagnostic Value in Colorectal Tumorous Lesions. Front. Mol. Biosci. 2021, 8, 706754.
- Chen, Z.; Zhao, G.; Wang, K.; Wang, X.; Ma, Y.; Xiong, S.; Zheng, M.; Fei, S. Blood leukocytes methylation levels analysis indicate methylated plasma test is a promising tool for colorectal cancer early detection. J. Cancer 2021, 12, 3678–3685.
- Zhang, W.; Yang, C.; Wang, S.; Xiang, Z.; Dou, R.; Lin, Z.; Zheng, J.; Xiong, B. SDC2 and TFPI2 Methylation in Stool Samples as an Integrated Biomarker for Early Detection of Colorectal Cancer. Cancer Manag. Res. 2021, 13, 3601–3617.
- Müller, H.M.; Oberwalder, M.; Fiegl, H.; Morandell, M.; Goebel, G.; Zitt, M.; Mühlthaler, M.; Öfner, D.; Margreiter, R.; Widschwendter, M. Methylation changes in faecal DNA: A marker for colorectal cancer screening? Lancet 2004, 363, 1283–1285.
- Shirahata, A.; Hibi, K. Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer. Res. 2014, 34, 4121–4125.
- Shirahata, A.; Sakuraba, K.; Goto, T.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; Hibi, K. Detection of vimentin (VIM) methylation in the serum of colorectal cancer patients. Anticancer. Res. 2010, 30, 5015–5018.
- Yi, J.M.; Dhir, M.; Guzzetta, A.A.; Iacobuzio-Donahue, C.A.; Heo, K.; Yang, K.M.; Suzuki, H.; Toyota, M.; Kim, H.M.; Ahuja, N. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2012, 33, 363–372.
- Nassar, F.J.; Msheik, Z.S.; Nasr, R.R.; Temraz, S.N. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin. Epigenet. 2021, 13, 111.
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394.
- Agarwal, A.; Balic, M.; El-Ashry, D.; Cote, R.J. Circulating Tumor Cells: Strategies for Capture, Analyses, and Propagation. Cancer J. 2018, 24, 70–77.
- Li, H.; Courtois, E.T.; Sengupta, D.; Tan, Y.; Chen, K.H.; Goh, J.J.L.; Kong, S.L.; Chua, C.; Hon, L.K.; Tan, W.S.; et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017, 49, 708–718.
- Bian, S.; Hou, Y.; Zhou, X.; Li, X.; Yong, J.; Wang, Y.; Wang, W.; Yan, J.; Hu, B.; Guo, H.; et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 2018, 362, 1060–1063.
- Denis, J.A.; Patroni, A.; Guillerm, E.; Pépin, D.; Benali-Furet, N.; Wechsler, J.; Manceau, G.; Bernard, M.; Coulet, F.; Larsen, A.K.; et al. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol. Oncol. 2016, 10, 1221–1231.
- Therkildsen, C.; Bergmann, T.K.; Henrichsen-Schnack, T.; Ladelund, S.; Nilbert, M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014, 53, 852–864.
- Nguyen, T.T.; Ung, T.T.; Li, S.; Sah, D.K.; Park, S.Y.; Lian, S.; Jung, Y.D. Lithocholic Acid Induces miR21, Promoting PTEN Inhibition via STAT3 and ERK-1/2 Signaling in Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 10209.
- Durán-Vinet, B.; Araya-Castro, K.; Calderón, J.; Vergara, L.; Weber, H.; Retamales, J.; Araya-Castro, P.; Leal-Rojas, P. CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review. Cancers 2021, 13, 4640.
- Gungormez, C.; Gumushan Aktas, H.; Dilsiz, N.; Borazan, E. Novel miRNAs as potential biomarkers in stage II colon cancer: Microarray analysis. Mol. Biol. Rep. 2019, 46, 4175–4183.
- Nagel, R.; le Sage, C.; Diosdado, B.; van der Waal, M.; Vrielink, J.A.O.; Bolijn, A.; Meijer, G.A.; Agami, R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008, 68, 5795–5802.
- Yang, I.-P.; Tsai, H.-L.; Miao, Z.-F.; Huang, C.-W.; Kuo, C.-H.; Wu, J.-Y.; Wang, W.-M.; Juo, S.-H.H.; Wang, J.-Y. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J. Transl. Med. 2016, 14, 108.
- Shi, Y.; Liu, Z. Serum miR-92a-1 is a novel diagnostic biomarker for colorectal cancer. J. Cell. Mol. Med. 2020, 24, 8363–8367.
- Sun, Y.; Yang, B.; Lin, M.; Yu, H.; Chen, H.; Zhang, Z. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019, 24, 299–305.
- Shen, X.; Xue, Y.; Cong, H.; Wang, X.; Fan, Z.; Cui, X.; Ju, S. Circulating lncRNA DANCR as a potential auxillary biomarker for the diagnosis and prognostic prediction of colorectal cancer. Biosci. Rep. 2020, 40, BSR20191481.
- Wang, Y.; Zhang, D.; Zhang, C.; Sun, Y. The Diagnostic and Prognostic Value of Serum lncRNA NEAT1 in Colorectal Cancer. Cancer Manag. Res. 2020, 12, 10985–10992.
- Xie, Y.; Li, J.; Li, P.; Li, N.; Zhang, Y.; Binang, H.; Zhao, Y.; Duan, W.; Chen, Y.; Wang, Y.; et al. RNA-Seq Profiling of Serum Exosomal Circular RNAs Reveals Circ-PNN as a Potential Biomarker for Human Colorectal Cancer. Front. Oncol. 2020, 10, 982.
- Chen, W.; Gao, C.; Liu, Y.; Wen, Y.; Hong, X.; Huang, Z. Bioinformatics Analysis of Prognostic miRNA Signature and Potential Critical Genes in Colon Cancer. Front. Genet. 2020, 11, 478.
- Hauptman, N.; Jevšinek Skok, D.; Spasovska, E.; Boštjančič, E.; Glavač, D. Genes CEP55, FOXD3, FOXF2, GNAO1, GRIA4, and KCNA5 as potential diagnostic biomarkers in colorectal cancer. BMC Med. Genom. 2019, 12, 54.
- Jung, Y.; Lee, S.; Choi, H.-S.; Kim, S.-N.; Lee, E.; Shin, Y.; Seo, J.; Kim, B.; Jung, Y.; Kim, W.; et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin. Cancer Res. 2011, 17, 700–709.
- Lin, K.; Zhu, X.; Luo, C.; Bu, F.; Zhu, J.; Zhu, Z. Data mining combined with experiments to validate CEP55 as a prognostic biomarker in colorectal cancer. Immun. Inflamm. Dis. 2021, 9, 167–182.
- Piran, M.; Sepahi, N.; Moattari, A.; Rahimi, A.; Ghanbariasad, A. Systems Biomedicine of Primary and Metastatic Colorectal Cancer Reveals Potential Therapeutic Targets. Front. Oncol. 2021, 11, 597536.
- Song, G.; Xu, S.; Zhang, H.; Wang, Y.; Xiao, C.; Jiang, T.; Wu, L.; Zhang, T.; Sun, X.; Zhong, L.; et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J. Exp. Clin. Cancer Res. 2016, 35, 148.
- Sun, G.; Li, Y.; Peng, Y.; Lu, D.; Zhang, F.; Cui, X.; Zhang, Q.; Li, Z. Identification of differentially expressed genes and biological characteristics of colorectal cancer by integrated bioinformatics analysis. J. Cell. Physiol. 2019, 234, 15215–15224.
- Yu, M.H.; Luo, Y.; Qin, S.L.; Wang, Z.S.; Mu, Y.F.; Zhong, M. Up-regulated CKS2 promotes tumor progression and predicts a poor prognosis in human colorectal cancer. Am. J. Cancer Res. 2015, 5, 2708–2718.
- Liu, J.; Li, H.; Sun, L.; Shen, S.; Zhou, Q.; Yuan, Y.; Xing, C. Epigenetic Alternations of MicroRNAs and DNA Methylation Contribute to Liver Metastasis of Colorectal Cancer. Dig. Dis. Sci. 2019, 64, 1523–1534.
- Liu, C.; Pan, Z.; Chen, Q.; Chen, Z.; Liu, W.; Wu, L.; Jiang, M.; Lin, W.; Zhang, Y.; Lin, W.; et al. Pharmacological targeting PTK6 inhibits the JAK2/STAT3 sustained stemness and reverses chemoresistance of colorectal cancer. J. Exp. Clin. Cancer Res. 2021, 40, 297.
- Li, R.; Hao, Y.; Wang, Q.; Meng, Y.; Wu, K.; Liu, C.; Xu, L.; Liu, Z.; Zhao, L. ECHS1, an interacting protein of LASP1, induces sphingolipid-metabolism imbalance to promote colorectal cancer progression by regulating ceramide glycosylation. Cell Death Dis. 2021, 12, 911.
- Yang, G.; Huang, L.; Jia, H.; Aikemu, B.; Zhang, S.; Shao, Y.; Hong, H.; Yesseyeva, G.; Wang, C.; Li, S.; et al. NDRG1 enhances the sensitivity of cetuximab by modulating EGFR trafficking in colorectal cancer. Oncogene 2021, 40, 5993–6006.
- Coss, A.; Tosetto, M.; Fox, E.J.; Sapetto-Rebow, B.; Gorman, S.; Kennedy, B.N.; Lloyd, A.T.; Hyland, J.M.; O’Donoghue, D.P.; Sheahan, K.; et al. Increased topoisomerase IIalpha expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis. Cancer Lett. 2009, 276, 228–238.
- Qu, X.; Sandmann, T.; Frierson, H.; Fu, L.; Fuentes, E.; Walter, K.; Okrah, K.; Rumpel, C.; Moskaluk, C.; Lu, S.; et al. Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene 2016, 35, 6403–6415.
- Dong, B.; Chai, M.; Chen, H.; Feng, Q.; Jin, R.; Hu, S. Screening and verifying key genes with poor prognosis in colon cancer through bioinformatics analysis. Transl. Cancer Res. 2020, 9, 6720–6732.
- Yu, D.; Sun, J.; Weng, Y.; Luo, L.; Sheng, J.; Xu, Z. Serum angiogenin as a potential biomarker for early detection of colorectal adenomas and colorectal cancer. Anti-Cancer Drugs 2021, 32, 703–708.




