
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yoshihisa Matsumoto | -- | 4018 | 2022-04-30 14:12:40 | | | |
| 2 | Peter Tang | -1832 word(s) | 2186 | 2022-05-05 11:39:48 | | |
Video Upload Options
DNA-dependent protein kinase (DNA-PK), which is composed of a DNA-PK catalytic subunit (DNA-PKcs) and Ku80-Ku70 heterodimer, acts as the molecular sensor for DSB and plays a pivotal role in DSB repair through non-homologous end joining (NHEJ). Cells deficient for DNA-PKcs show hypersensitivity to IR and several DNA-damaging agents. Cellular sensitivity to IR and DNA-damaging agents can be augmented by the inhibition of DNA-PK. A number of small molecules that inhibit DNA-PK have been developed.
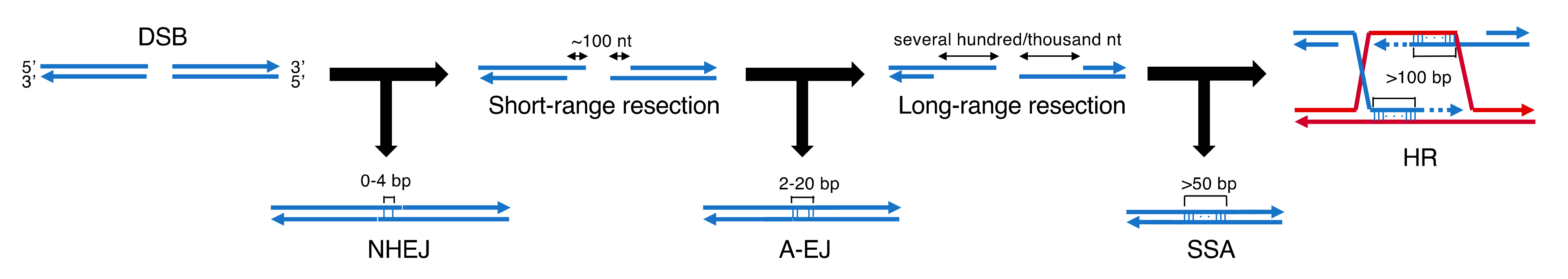
1. DNA double-strand break repair
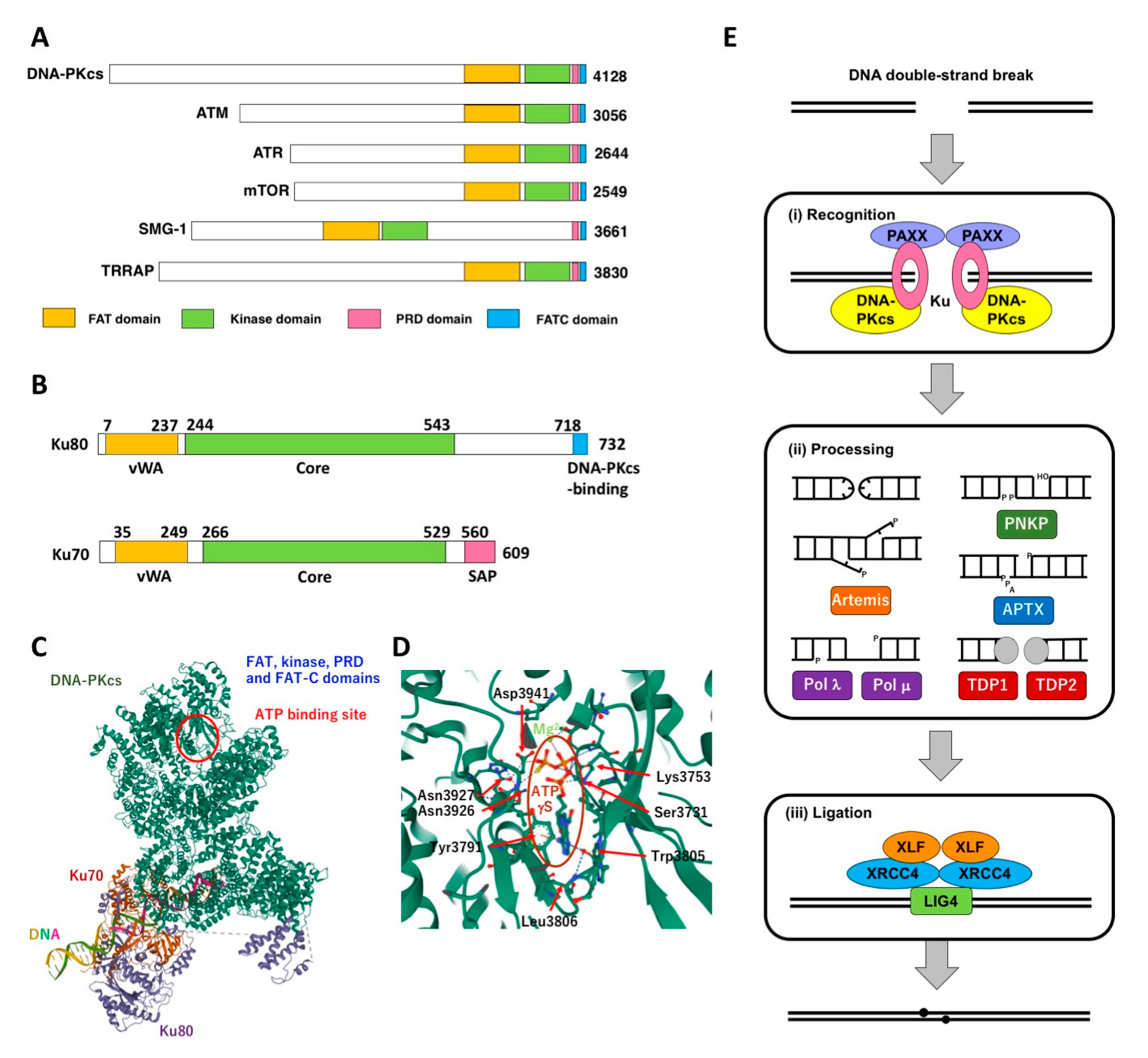
2. DNA-PK and Its Role NHEJ
|
Substrates |
Function |
Substrates |
Function |
|---|---|---|---|
|
[DNA Repair and Damage Signaling] |
[Transcription] |
||
|
(NHEJ) |
RNA polymerase II |
Transcription (general) |
|
|
DNA-dependent protein kinase catalytic subunit (DNA-PKcs) |
DNA-PK complex |
||
|
TATA box-binding protein (TBP) |
|||
|
Ku autoantigen 80kDa subunit (Ku80) |
p53 |
Transcription (specific) |
|
|
Specificity protein 1 (Sp1) |
|||
|
Ku autoantigen 70 kDa subunit (Ku70) |
c-Jun |
||
|
c-Fos |
|||
|
DNA ligase IV (LIG4) |
Ligation complex |
c-Myc |
|
|
X-ray repair cross-complementing group 4 (XRCC4) |
Octamer-binding factor 1 (Oct-1) |
||
|
XRCC4-like factor (XLF) |
Serum response factor (SRF) |
||
|
Artemis |
Nuclease |
[RNA metabolism] |
|
|
Polynucleotide kinase phosphatase (PNKP) |
Kinase, phosphatase |
Nuclear DNA helicase II (NDHII) |
Transcription and RNA processing |
|
Werner syndrome protein (WRN) |
Helicase, nuclease |
Heterogeneous nuclear ribonucleoprotein A1 (hnRNP-A1) |
RNA splicing |
|
(Other DNA repair and damage signaling pathways) |
|||
|
Ataxia telangiectasia mutated (ATM) |
Protein kinase; HR and cell cycle checkpoint |
Heterogeneous nuclear ribonucleoprotein U (hnRNP-U) |
|
|
Replication protein A 2 (RPA2) |
Single-stranded DNA binding; HR and DNA replication |
Fused in sarcoma (FUS) |
RNA binding |
|
Poly(ADP-ribose) polymerase 1 (PARP1) |
Single-strand break repair |
[Signaling] |
|
|
Excision repair cross complementing 1 (ERCC1) |
Nuclease component; nucleotide excision repair |
Akt1 |
Protein kinase |
|
Akt2 |
Protein kinase |
||
|
[DNA replication] |
Sty1/Spc1-interacting protein 1 (Sin1) |
Protein kinase regulator |
|
|
DNA ligase I (LIG1) |
Ligation |
||
|
Minichromosome maintenance 3 (MCM3) |
Initiation of replication |
[Organelle, cytoskeleton] |
|
|
[Nucleosome and chromatin structure] |
Golgi phosphoprotein 3 (GOLPH3) |
Linking Golgi membrane to cytoskeleton |
|
|
Histone H2AX |
Core histone component; recruitment of DSB repair proteins |
Vimentin |
Intermediate filament |
|
Histone H1 |
Linker histone |
Tau |
Microtubule regulation |
|
High mobility group 1 (HMG1) |
Maintenance and regulation of chromatin structure |
[Protein maintenance] |
|
|
High mobility group 2 (HMG2) |
Heat shock protein 90 alpha (HSP90a) |
Protein chaperone |
|
|
C1D |
Valosin-containing protein (VCP) |
AAA+ ATPase |
|
|
Topoisomerase I |
Regulation of topological status of DNA |
[Metabolism] |
|
|
Topoisomerase II |
Fumarate hydratase (FH) |
Production of L-malate from fumarate; regulation of NHEJ |
|
|
Nuclear orphan receptor 4A2 (NR4A2) |
Chromatin regulation; regulation of NHEJ |
||
|
Pituitary tumor-transforming gene (PTTG) |
Regulation of chromosome segregation |
||
References
- United Nations Scientific Committee on the Effects of Atomic Radiation. Report of the United Nations Scientific Committee on the Effects of Atomic Radiation 2010 Fifty-Seventh Session, Includes Scientific Report: Summary of Low-Dose Radiation Effects on Health; United Nations Scientific Committee on the Effects of Atomic Radiation: Vienna, Austria, 2010.
- Zhao, B.; Rothenberg, E.; Ramsden, D.A.; Lieber, M.R. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol. 2020, 21, 765–781.
- Yilmaz, D.; Furst, A.; Meaburn, K.; Lezaja, A.; Wen, Y.; Altmeyer, M.; Reina-San-Martin, B.; Soutoglou, E. Activation of homologous recombination in G1 preserves centromeric integrity. Nature 2021, 600, 748–753.
- Dvir, A.; Stein, L.Y.; Calore, B.L.; Dynan, W.S. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem. 1993, 268, 10440–10447.
- Gottlieb, T.M.; Jackson, S.P. The DNA-dependent protein kinase: Requirement for DNA ends and association with Ku antigen. Cell 1993, 72, 131–142.
- Hartley, K.; Gell, D.; Smith, C.; Zhang, H.; Divecha, N.; Connelly, M.; Admon, A.; Lees-Miller, S.; Anderson, C.; Jackson, S. DNA-dependent protein kinase catalytic subunit: A relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 1995, 82, 849–856.
- Savitsky, K.; Bar-Shira, A.; Gilad, S.; Rotman, G.; Ziv, Y.; Vanagaite, L.; Tagle, D.A.; Smith, S.; Uziel, T.; Sfez, S.; et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995, 268, 1749–1753.
- Bentley, N.J.; Holtzman, D.A.; Flaggs, G.; Keegan, K.S.; DeMaggio, A.; Ford, J.C.; Hoekstra, M.; Carr, A.M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996, 15, 6641–6651.
- Cimprich, K.A.; Shin, T.B.; Keith, C.T.; Schleiber, S.L. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. USA 1996, 93, 2850–2855.
- Girard, P.M.; Riballo, E.; Begg, A.C.; Waugh, A.; Jeggo, P.A. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene 2002, 21, 4191–4199.
- Uziel, T.; Lerenthal, Y.; Moyal, L.; Andegeko, Y.; Mittelman, L.; Shiloh, Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003, 22, 5612–5621.
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548.
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758.
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a Rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43.
- Denning, G.; Jamieson, L.; Maquat, L.E.; Thompson, E.A.; Fields, A.P. Cloning of a novel phosphatidylinositol kinase-related kinase: Characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem. 2001, 276, 22709–22714.
- Yamashita, A.; Ohnishi, T.; Kashima, I.; Taya, Y.; Ohno, S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001, 15, 2215–2228.
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998, 94, 363–374.
- Alemi, F.; Sadigh, A.R.; Malakoti, F.; Elhaei, Y.; Ghaffari, S.H.; Maleki, M.; Asemi, Z.; Yousefi, B.; Targhazeh, N.; Majidinia, M. Molecular mechanisms involved in DNA repair in human cancers: An overview of PI3k/Akt signaling and PIKKs crosstalk. J. Cell Phisiol. 2022, 237, 313–328.
- Huang, T.T.; Lampert, E.J.; Coots, C.; Lee, J.M. Targeting the PI3K pathway and DNA damage response as a therapeutic strategy in ovarian cancer. Cancer Treat. Rev. 2020, 86, 102021.
- Wanigasooriya, K.; Tyler, R.; Barros-Silva, J.D.; Sinha, Y.; Ismail, T.; Beggs, A.D. Radiosensitising Cancer Using Phosphatidylinositol-3-Kinase (PI3K), Protein Kinase B (AKT) or Mammalian Target of Rapamycin (mTOR) Inhibitors. Cancers (Basel) 2020, 12, 1278.
- Gewandter, J.S.; Bambara, R.A.; O’Reilly, M.A. The RNA surveillance protein SMG1 activates p53 in response to DNA double-strand breaks but not exogenously oxidized mRNA. Cell Cycle 2011, 10, 2561–2567.
- Gubanova, E.; Issaeva, N.; Gokturk, C.; Djureinovic, T.; Helleday, T. SMG-1 suppresses CDK2 and tumor growth by regulating both the p53 and Cdc25A signaling pathways. Cell Cycle 2013, 12, 3770–3780.
- Zhang, C.; Mi, J.; Deng, Y.; Deng, Z.; Long, D.; Liu, Z. DNMT1 Enhances the Radiosensitivity of HPV-Positive Head and Neck Squamous Cell Carcinomas via Downregulating SMG1. Onco. Targets Ther. 2020, 13, 4201–4211.
- Long, D.; Xu, L.; Deng, Z.; Guo, D.; Zhang, Y.; Liu, Z.; Zhang, C. HPV16 E6 enhances the radiosensitivity in HPV-positive human head and neck squamous cell carcinoma by regulating the miR-27a-3p/SMG1 axis. Infect. Agent Cancer 2021, 16, 56.
- Chen, Y.H.; Wei, M.F.; Wang, C.W.; Lee, H.W.; Pan, S.L.; Gao, M.; Kuo, S.H.; Cheng, A.L.; Teng, C.M. Dual phosphoinositide 3-kinase/mammalian target of rapamycin inhibitor is an effective radiosensitizer for colorectal cancer. Cancer Lett. 2015, 357, 582–590.
- Del Peso, L.; González-García, M.; Page, C.; Herrera, R.; Nuñez, G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 1997, 278, 687–689.
- Datta, S.R.; Dudek, H.; Tao, X.; Masters, S.; Fu, H.; Gotoh, Y.; Greenberg, M.E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997, 91, 231–241.
- Zhou, B.P.; Liao, Y.; Xia, W.; Zou, Y.; Spohn, B.; Hung, M.C. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 2001, 3, 973–982.
- Oeck, S.; Al-Refae, K.; Riffkin, H.; Wiel, G.; Handrick, R.; Klein, D.; Iliakis, G.; Jendrossek, V. Activating Akt1 mutations alter DNA double strand break repair and radiosensitivity. Sci. Rep. 2017, 7, 42700.
- Mueck, K.; Rebholz, S.; Harati, M.D.; Rodemann, H.P.; Toulany, M. Akt1 Stimulates Homologous Recombination Repair of DNA Double-Strand Breaks in a Rad51-Dependent Manner. Int. J. Mol. Sci. 2017, 18, 2473.
- Bozulic, L.; Surucu, B.; Hynx, D.; Hemmings, B.A. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol. Cell 2008, 30, 203–213.
- Liu, L.; Dai, X.; Yin, S.; Liu, P.; Hill, E.G.; Wei, W.; Gan, W. DNA-PK promotes activation of the survival kinase AKT in response to DNA damage through an mTORC2-ECT2 pathway. Sci. Signal 2022, 15, eabh2290.
- Matsumoto, Y.; Asa, A.D.D.C.; Modak, C.; Shimada, M. DNA-dependent protein kinase catalytic subunit: The sensor for DNA double-strand breaks structurally and functionally related to Ataxia telangiectasia mutated. Genes 2021, 12, 1143.
- Matsumoto, Y.; Sharma, M.K. DNA-dependent protein kinase in DNA damage response: Three decades and beyond. J. Radiat. Cancer Res. 2020, 11, 123–134.
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, 431–437.
- Yaneva, M.; Wen, J.; Ayala, A.; Cook, R. cDNA-derived amino acid sequence of the 86-kDa subunit of the Ku antigen. J. Biol. Chem. 1989, 264, 13407–13411.
- Reeves, W.H.; Sthoeger, Z.M. Molecular cloning of cDNA encoding the p70 (Ku) lupus autoantigen. J. Biol. Chem. 1989, 264, 5047–5052.
- Mimori, T.; Hardin, J.A. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 1986, 261, 10375–10379.
- Falck, J.; Coates, J.; Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434, 605–611.
- Walker, J.R.; Corpina, R.A.; Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implication for double-strand break repair. Nature 2001, 412, 607–614.
- Chen, X.; Xu, X.; Chen, Y.; Cheung, J.C.; Wang, H.; Jiang, J.; de Val, N.; Fox, T.; Gellert, M.; Yang, W. Structure of an activated DNA-PK and its implications for NHEJ. Mol. Cell 2021, 81, 801–810.
- Liang, S.; Thomas, S.E.; Chaplin, A.K.; Hardwick, S.W.; Chirgadze, D.Y.; Blundell, T.L. Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs. Nature 2022, 601, 643–648.
- Taccioli, G.E.; Gottlieb, T.M.; Blunt, T.; Priestley, A.; Demengeot, J.; Mizuta, R.; Lehmann, A.R.; Alt, F.W.; Jackson, S.P.; Jeggo, P.A. Ku80: Product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 1994, 265, 1442–1445.
- Smider, V.; Rathmell, W.K.; Lieber, M.R.; Chu, G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science 1994, 266, 288–291.
- Blunt, T.; Finnie, N.J.; Taccioli, G.E.; Smith, G.C.; Demengeot, J.; Gottlieb, T.M.; Mizuta, R.; Varghese, A.J.; Alt, F.W.; Jeggo, P.A. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 1995, 80, 813–823.
- Kirchgessner, C.U.; Patil, C.K.; Evans, J.W.; Cuomo, C.A.; Fried, L.M.; Carter, T.; Oettinger, M.A.; Brown, J.M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science 1995, 267, 1178–1183.
- Peterson, S.R.; Kurimasa, A.; Oshimura, M.; Dynan, W.S.; Bradbury, E.M.; Chen, D.J. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc. Natl. Acad. Sci. USA 1995, 92, 3171–3174.
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 2015, 347, 185–188.
- Xing, M.; Yang, M.; Huo, W.; Feng, F.; Wei, L.; Jiang, W.; Ning, S.; Yan, Z.; Li, W.; Wang, Q.; et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015, 6, 6233.
- Craxton, A.; Somers, J.; Munnur, D.; Jukes-Jones, R.; Cain, K.; Malewicz, M. XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death Diff. 2015, 22, 890–897.
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001, 105, 177–186.
- Ma, Y.; Pannicke, U.; Schwarz, K.; Lieber, M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 2002, 108, 781–794.
- Li, Z.; Otevrel, T.; Gao, Y.; Cheng, H.; Seed, B.; Stamato, T.; Taccioli, G.E.; Alt, F.W. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 1995, 83, 1079–1089.
- Critchlow, S.; Bowater, R.; Jackson, S. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997, 7, 588–598.
- Grawunder, U.; Wilm, M.; Wu, X.; Kulesza, P.; Wilson, T.; Mann, M.; Leiber, M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 1997, 388, 492–495.
- Buck, D.; Malivert, L.; de Chasseval, R.; Barraud, A.; Fondanèche, M.C.; Sanal, O.; Plebani, A.; Stéphan, J.L.; Hufnagel, M.; le Deist, F.; et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 2006, 124, 287–299.
- Ahnesorg, P.; Smith, P.; Jackson, S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 2006, 124, 301–313.
- Hammel, M.; Yu, Y.; Fang, S.; Lees-Miller, S.P.; Tainer, J.A. XLF regulates filament architecture of the XRCC4·ligase IV complex. Structure 2010, 18, 1431–1442.
- Kienker, L.J.; Shin, E.K.; Meek, K. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 2000, 28, 2752–2761.
- Kurimasa, A.; Kumano, S.; Boubnov, N.; Story, M.; Tung, C.; Peterson, S.; Chen, D.J. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell Biol. 1999, 19, 3877–3884.




