Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stefania Albrizio | -- | 1332 | 2022-04-28 12:19:32 | | | |
| 2 | Catherine Yang | -141 word(s) | 1191 | 2022-04-29 03:21:36 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Albrizio, S.; Schiano, M.; , .; Varra, M. Aptamer-Based Biosensors for Bisphenol A in Foodstuffs. Encyclopedia. Available online: https://encyclopedia.pub/entry/22434 (accessed on 07 February 2026).
Albrizio S, Schiano M, , Varra M. Aptamer-Based Biosensors for Bisphenol A in Foodstuffs. Encyclopedia. Available at: https://encyclopedia.pub/entry/22434. Accessed February 07, 2026.
Albrizio, Stefania, Marica Schiano, , Michela Varra. "Aptamer-Based Biosensors for Bisphenol A in Foodstuffs" Encyclopedia, https://encyclopedia.pub/entry/22434 (accessed February 07, 2026).
Albrizio, S., Schiano, M., , ., & Varra, M. (2022, April 28). Aptamer-Based Biosensors for Bisphenol A in Foodstuffs. In Encyclopedia. https://encyclopedia.pub/entry/22434
Albrizio, Stefania, et al. "Aptamer-Based Biosensors for Bisphenol A in Foodstuffs." Encyclopedia. Web. 28 April, 2022.
Copy Citation
Bisphenol A (BPA) is a synthetic compound utilized to manufacture plastics for Food Contact Materials (FCMs) or resins for the inside of food containers. Since it was recognized as an Endocrine-Disrupting Chemical (EDC), its implications in pathologies, such as cancer, obesity, diabetes, immune system alterations, and developmental and mental disorders, have been widely documented. Diet is considered the main source of exposure for humans to BPA. Consequently, continuous monitoring of the levels of BPA in foods is necessary to assess the risk associated with its consumption in one’s diet.
bisphenols
Endocrine-Disrupting Chemicals (EDCs)
biosensors
aptasensors
1. Introduction
Bisphenol A (BPA, Figure 1) is a synthetic chemical with a long history of use. Since 1950, it has been utilized to produce epoxy resins, and it has also been widely applied in the plastic industry as a monomer to synthesize polycarbonate polymers or as an additive in the synthesis of other kinds of plastics, such as polyvinyl chloride (PVC) [1]. BPA is mainly used in manufacturing Food Contact Materials (FCMs), such as storage containers, plastic water bottles, food packaging, and the inner coatings of food cans [2], but it is also contained in electronics equipment, children’s toys, dental sealants, thermal paper, and flame retardants.
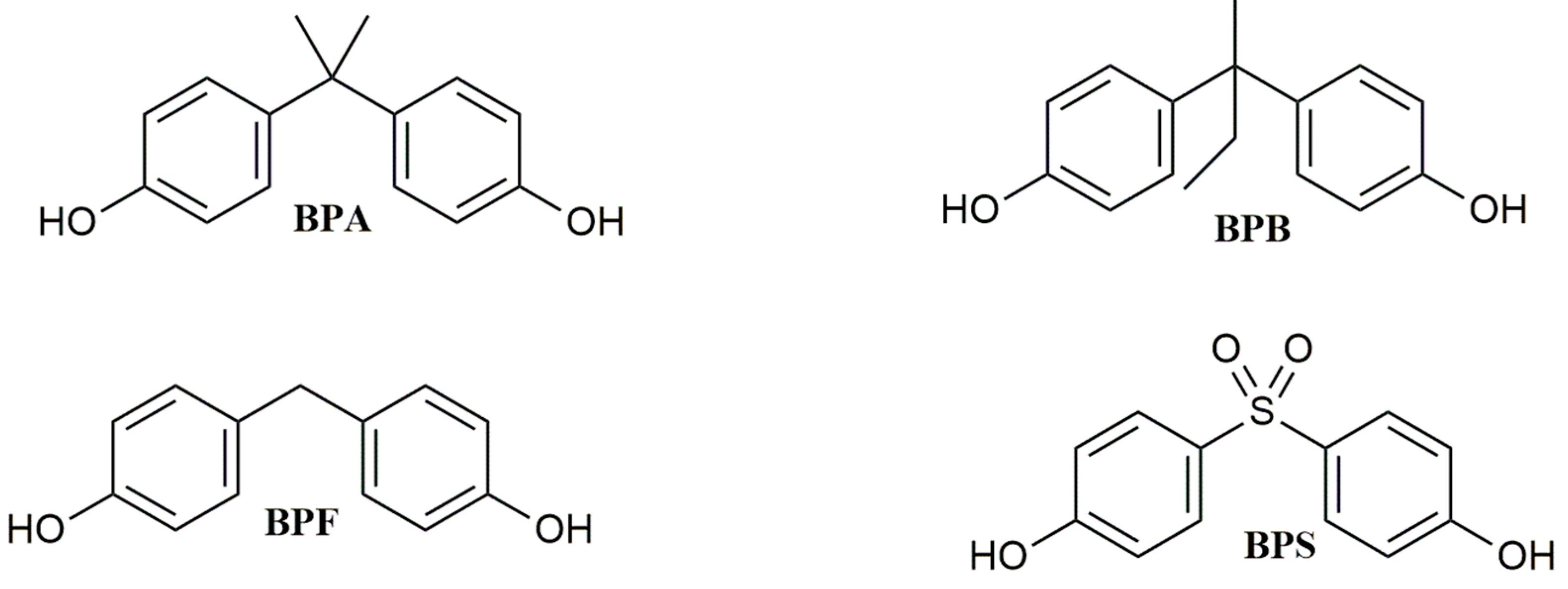
Figure 1. Chemical structure of BPA and its analogs.
2. BPA Aptamers: Sequence and Three-Dimensional Structure
Aptamers are sequence-specific single-stranded DNA or RNA oligonucleotides (12–200 nts) that are able to bind to a selected target with high affinity and selectivity. Many studies have shown that aptamers recognize their specific targets employing precise secondary or tertiary structures (duplex, G-quadruplex, stem-loop, etc.). Aptamers are selected by applying the Systematic Evolution of Ligands by EXponential enrichment (SELEX protocol) [3]. This procedure consists of an iterative selection and amplification process to identify, among a large pool of randomly generated single-stranded DNA/RNA sequences, the DNA/RNA molecules able to selectively bind to the immobilized target. After its development, SELEX protocol has been applied to obtain aptamers for different type of targets, varying from small molecules to proteins or also cells [4]. In analytical chemistry, aptamers have been successful applied as key elements to build up sensors able to capture and quali-quantitatively analyze a specific substance in a certain matrix. A general scheme of the principal steps required to build up an aptasensor is depicted in Figure 2.
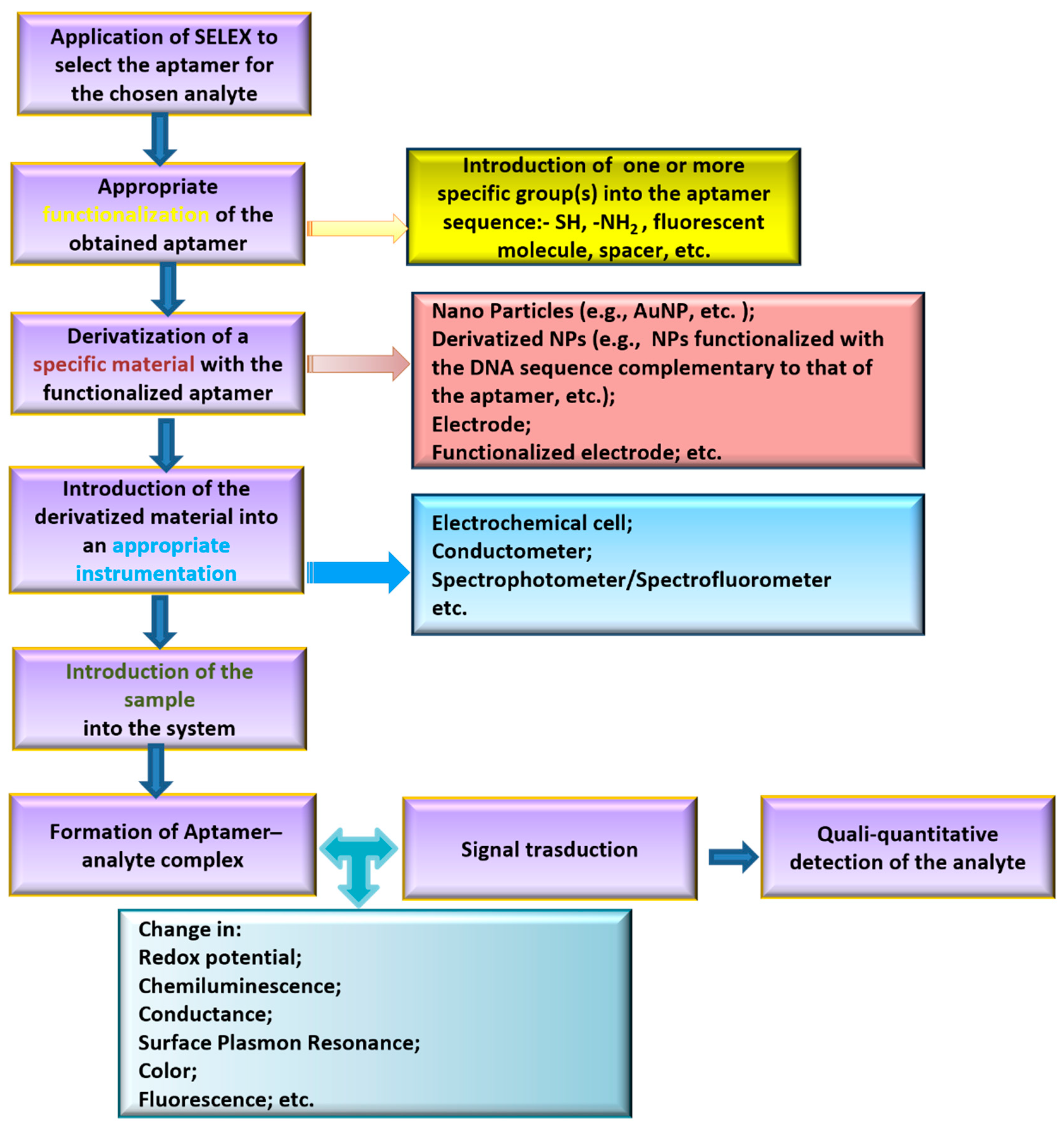
Figure 2. Scheme of the general strategy for aptasensor development.
Since the first anti-BPA aptamers were selected by Jo et al. [5], many efforts to develop sensitive BPA aptasensors for their use in different matrices have been made. With its high affinity and selectivity, BPA-Apt-2 (Table 1) is the main adopted sequence in the building up of aptasensors, and despite its 63 nt length, no truncated sequence has been developed until 2017 from the initial aptamer. Starting from the predicted secondary structure of the aptamer, Lee et al. [6] identified that BPA-Apt-3 (Table 1) was able to bind to BPA with increased affinity and selectivity compared to BPA-Apt-2.
Table 1. Aptamers against BPA used in biosensors for BPA detection and quantification in foods.
| Aptamer Binding BPA | Reference | ||||
|---|---|---|---|---|---|
| Acronym | Length | Sequence | Kd [nM] * | ||
| BPA-Apt-1 | 60 nts | CCGCCGTTGGTGTGGTGGGCCTAGGGCCGGCGGCGCACAGCTGTTATAGACGCCTCCAGC | not reported | [5] | Table 1, group 2; ID #6 * |
| BPA-Apt-2 | 63 nts | CCGGTGGGTGGTCAGGTGGGATAGCGTTCCGCGTATGGCCCAGCGCATCACGGGTTCGCACCA | 8.3 ** | [5] | Table 1, group 7; ID #3 * |
| BPA-Apt-3 | 24 nts | TTTTTTTTTTGGATAGCGGGTTCC | not reported | [6] | Truncated BPA-Apt-2 |
| BPA-Apt-4 | 38 nts | TGGGTGGTCAGGTGGGATAGCGTTCCGCGTATGGCCCA | 13.17 ± 1.02 *** | [7] | Truncated BPA-Apt-2 |
| BPA-Apt-5 | 12 nts | GGATAGCGTTCC | 27.05 ± 2.08 *** | [7] | Truncated BPA-Apt-2 |
| BPA-Apt-6 | 23 nts | TTTTTTTTTTCCGGTGGGTGGAA | 1190.61 ± 66.05 *** | [6] | Truncated BPA-Apt-2 |
The comparison between the three-dimensional structure predicted for BPA-Apt-3 and that of BPA-Apt-5 shows the presence of the same stem, formed by the 5’-GGA and 3’-TCC, and a differently long loop containing a common AGCGT sequence in both the folded aptamers. These two works confirm that SELEX alone is unsatisfactory for obtaining an efficient and cost-effective aptamer. Therefore, further studies are necessary to identify the exact binding mode of BPA to its aptamers and/or to the shortened aptamer derivatives as well as the secondary structures adopted in solution by the aptamer(s) alone and in the BPA–aptamer complex(es). Indeed, only a few studies reported the molecular modeling of aptamers and their BPA complexes [6][7], whereas in most cases, the stem-loop aptamer structures have been derived based on predictive DNA programs, and only in few cases they were confirmed by circular dichroism analysis. Notably, some authors mentioned the ability of BPA to induce the folding of BPA-Apt-2 (Table 1) into G-quadruplex(s) [9][10][11][12], although this hypothesis requires further experimental evidence.
3. Aptamer-Based Biosensor Applications Tested for the Determination of BPA in Real Food Samples
So far, aptamer-based biosensors have not been applied to an extensive monitoring of the levels of BPA in food samples. However, several authors reported the designs of various kinds of aptasensors to be specifically used in the detection of BPA in food samples. After their construction, all the aptasensors were tested to verify their feasibility in the analysis of real samples. All foods used in the applicability tests were liquids, mostly tap or mineralized water [7][11][13][14][15][16][17][18][19][20][21][22][23], followed by milk and fruit juices [9][15][21][22][24][25] and red wine samples [26]. Either no treatment (water samples) or just a quick pre-treatment of samples was necessary. Fruit juices were filtered to remove any residues of solid particles. Milk samples required a slightly time-consuming pre-treatment to eliminate fats by centrifugation and to eliminate proteins by precipitation and further centrifugation. Mirzajani et al. [27] analyzed two different brands of canned food (green beans and sweet peas), but they carried out the experiments only on the liquid portion of the cans. However, studies in the literature demonstrated that, when canned foods are analyzed, the BPA concentrations are higher in the solid portion than those detected in the liquid portion [28][29]. Especially in the case of canned seafoods, this should be considered in relation to the high probability of contamination due to the ingestion of microplastics by fish in the marine environment [30][31][32]. At last, Lee et al. [33] investigated the application of their colorimetric aptasensor directly on grains of commercially available steamed rice pre-treated with a solution of BPA.
The limit of detection (LOD) and recovery at different concentrations of BPA were the main performance parameters that the authors evaluated. In addition, the selectivity of each aptasensor was generally assessed vs. possible interferent substances or BPA analogs, including some compounds currently used as substitutes of BPA in plastic manufacturing. In several studies the reproducibility was also measured. Both selectivity and reproducibility proved to be The LODs, estimated mostly using BPA solutions at different concentrations, were generally very low. Jia et al. [7] compared the performance of their aptasensor to other more traditional methods. They showed that the LOD obtained by the aptasensor was generally lower, except for the case of GC–MS and electrochemical methods. Nonetheless, the authors concluded that their detection technique was preferable considering the undoubted advantages offered by the aptasensor, such as a low cost and a simple detection process in addition to a still-high sensitivity.
Generally, aptasensors tested in food samples showed good stability. Only Ye et al. found that their biosensor was not very stable when compared to the LC–MS/MS method, although they suggested that this problem could be overcome by carefully checking the temperature and buffering solution [21].
References
- Vogel, S. The Politics of Plastics: The Making and Unmaking of Bisphenol A Safety. Am. J. Public Health 2009, 99, 559–566.
- Almeida, S.; Raposo, A. Bisphenol A: Food Exposure and Impact on Human Health. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1503–1517.
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510.
- Kohlberger, M.; Gadermaier, G. SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnol. Appl. Biochem. 2021, 1–22.
- Jo, M.; Ahn, J.Y.; Lee, J.; Lee, S.; Hong, S.W.; Yoo, J.W.; Kang, J.; Dua, P.; Lee, D.; Hong, S.; et al. Development of Single-Stranded DNA Aptamers for Specific Bisphenol A Detection. Oligonucleotides 2011, 21, 85–91.
- Lee, E.H.; Lim, H.J.; Lee, S.D.; Son, A. Highly Sensitive Detection of Bisphenol A by NanoAptamer Assay with Truncated Aptamer. ACS Appl. Mater. Interfaces 2017, 9, 14889–14898.
- Jia, M.; Sha, J.; Li, Z.; Wang, W.; Zhang, H. High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem. 2020, 317, 126459.
- Ma, Y.; Liu, J.; Li, H. Diamond-based electrochemical aptasensor realizing a femtomolar detection limit of bisphenol A. Biosens. Bioelectron. 2017, 92, 21–25.
- Zhou, L.; Wang, J.; Li, D.; Li, Y. An electrochemical aptasensor based on gold nanoparticles dotted graphene modified glassy carbon electrode for label-free detection of bisphenol A in milk samples. Food Chem. 2014, 162, 34–40.
- He, M.Q.; Wang, K.; Wang, J.; Yu, Y.L.; He, R.H. A sensitive aptasensor based on molybdenum carbide nanotubes and label-free aptamer for detection of bisphenol A. Anal. Bioanal Chem. 2017, 409, 1797–1803.
- Qiao, Y.; Li, J.; Li, H.; Fang, H.; Fan, D.; Wang, W.A. A label-free photoelectrochemical aptasensor for bisphenol A based on surface plasmon resonance of gold nanoparticle-sensitized ZnO nanopencils. Biosens. Bioelectron. 2016, 1, 315–320.
- Tsekeli, T.R.; Tshwenya, L.; Sebokolodi, T.I.; Ndlovu, T.; Arotiba, O.A. An Electrochemical Aptamer Biosensor for Bisphenol A on a Carbon Nanofibre-silver Nanoparticle Immobilisation Platform. Electroanalysis 2021, 33, 2053–2061.
- Chung, C.; Jeon, J.; Yu, J.; Lee, C.; Choo, J. Surface-enhanced Raman scattering aptasensor for ultrasensitive trace analysis of bisphenol A. Biosens. Bioelectron. 2015, 64, 560–565.
- Zhu, Y.; Cai, Y.; Xu, L.; Zheng, L.; Wang, L.; Qi, L.; Xu, C. Building An Aptamer/Graphene Oxide FRET Biosensor for One-Step Detection of Bisphenol A. ACS Appl. Mater. Interfaces 2015, 7, 7492–7496.
- Deiminiat, B.; Rounaghi, G.H.; Arbab-Zavar, H.M.; Razavipanah, I. A novel electrochemical aptasensor based on f-MWCNTs/GNPs nanocomposite for label-free detection of bisphenol A. Sens. Actuators B Chem. 2017, 242, 158–166.
- Guo, X.; Wu, S.; Duan, N.; Wang, Z. Mn2+ -doped NaYF 4: Yb/Er upconversion nanoparticle-based electrochemiluminescent aptasensor for bisphenol A. Anal. Bioanal. Chem. 2016, 408, 3823–3831.
- Liu, Y.; Liu, Y.; Liu, B. A dual-signaling strategy for ultrasensitive detection of bisphenol A by aptamer-based electrochemical biosensor. J. Electroanal. Chem. 2016, 781, 265–271.
- Feng, J.; Xu, L.; Cui, G.; Wu, X.; Ma, W.; Kuang, H.; Xu, C. Building SERS-active heteroassemblies for ultrasensitive Bisphenol A detection. Biosens. Bioelectron. 2016, 81, 138–142.
- Zhu, Y.; Gu, X.; Jiang, F.; Jia, R.; Jin, M.; Chen, M.; Zhang, G. Ultrasensitive detection of Bisphenol A based on an aptasensor with DNA amplification. Food Agr. Immunol. 2018, 29, 1106–1115.
- Beiranvand, Z.S.; Abbasi, A.R.; Dehdashtian, S.; Karimi, Z.; Azadbakht, A. Aptamer-based electrochemical biosensor by using Au-Pt nanoparticles, carbon nanotubes and acriflavine platform. Anal. Biochem. 2017, 518, 34–45.
- Ye, S.; Ye, R.; Shi, Y.; Qiu, B.; Guo, L.; Huang, D.; Lin, Z.; Chen, G. Highly sensitive aptamer based on electrochemiluminescence biosensor for label-free detection of bisphenol A. Anal. Bioanal. Chem. 2017, 409, 7145–7151.
- Farahbakhsh, F.; Heydari-Bafrooei, E.; Ahmadi, M.; Hekmatara, S.H.; Sabet, M. A novel aptasensing method for detecting bisphenol A using the catalytic effect of the Fe3O4/Au nanoparticles on the reduction reaction of the silver ions. Food Chem. 2021, 355, 129666–129673.
- Peng, Y.; Liu, Y.; Zhang, X.; Zhou, J.; Xiong, E.; Li, X.; Chen, J. A novel electrochemical aptasensor for bisphenol A assay based on triple-signaling strategy. Biosens. Bioelectron. 2016, 79, 22–28.
- Ren, H.; An, Z.; Jang, C. Liquid crystal-based aptamer sensor for sensitive detection of bisphenol A. Microchem. J. 2019, 146, 1064–1071.
- Xu, Z.; Chen, Y.; Tang, Y.; Chen, M.; Chenand, W.; Cheng, Y. Aptamer-enhanced fluorescence determination of bisphenol A after magnetic solid-phase extraction using Fe3O4@SiO2@aptamer. Anal. Methods 2020, 12, 4479–4486.
- Shi, L.; Rong, X.; Wang, Y.; Dinga, S.; Tang, W. High-performance and versatile electrochemical aptasensor based on selfsupported nanoporous gold microelectrode and enzyme-induced signal. Biosens. Bioelectron. 2018, 102, 41–48.
- Mirzajani, H.; Cheng, C.; Wua, J.; Chen, J.; Edad, S.; Aghdamb, E.N.; Ghavifekr, H.B. A highly sensitive and specific capacitive aptasensor for rapid and label-free trace analysis of Bisphenol A (BPA) in canned foods. Biosens. Bioelectron. 2017, 89, 1059–1067.
- Fattore, M.; Russo, G.; Barbato, F.; Grumetto, L.; Albrizio, S. Monitoring of bisphenols in canned tuna from Italian markets. Food Chem. Toxicol. 2015, 83, 68–75.
- Noonan, G.; Ackerman, L.; Begley, T.H. Concentration of Bisphenol A in Highly Consumed Canned Foods on the U.S. Market. Food Chem. 2011, 59, 7178–7185.
- Liu, X.; Shi, H.; Xie, B.; Dionysiou, D.D.; Zhao, Y. Microplastics as Both a Sink and a Source of Bisphenol A in the Marine Environment. Environ. Sci. Technol. 2019, 53, 10188–10196.
- Barboza, L.; Cunha, S.C.; Monteiro, C.; Fernandes, J.O. Bisphenol A and its analogs in muscle and liver of fish from the North East Atlantic Ocean in relation to microplastic contamination. Exposure and risk to human consumers. J. Hazard. Mater. 2020, 393, 122419–122429.
- Bakir, A.; O’Connor, I.; Rowland, S.J.; Hendriks, A.J.; Thompson, R. Relative importance of microplastics as a pathway for the transfer of hydrophobic organic chemicals to marine life. Environ. Pollut. 2016, 219, 56–65.
- Lee, E.H.; Leeb, S.K.; Kim, M.J.; Lee, S.W. Simple and rapid detection of bisphenol A using a gold nanoparticle-based colorimetric aptasensor. Food Chem. 2019, 287, 205–213.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
29 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




