
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Abdullah Kaviani Rad | -- | 2232 | 2022-04-28 10:17:52 | | | |
| 2 | Catherine Yang | Meta information modification | 2232 | 2022-04-28 10:43:34 | | |
Video Upload Options
Excessive use of antibiotics in the healthcare sector and livestock farming has amplified antimicrobial resistance (AMR) as a major environmental threat in recent years. Abiotic stresses, including soil salinity and water pollutants, can affect AMR in soils, which in turn reduces the yield and quality of agricultural products.
1. Introduction
Soil microbial communities have manifold benefits for plants, including nutrient supply, synthesis of phytohormones, antagonistic activity against phytopathogens, and generation of signal molecules involved in microbe–plant interactions [1]. Although microbial communities are indicators of overall soil health, their resistance to adverse conditions for survival and competition is a considerable concern [2] owing to their potential to create ecosystem imbalances and disease emergence [3]. Hence, many studies have been carried out on microbial ecology in the environment [4], and most investigations concentrate on bioactive antimicrobial compounds in fertilizers, soil, and water resources [5]. Antibiotics, as one of the most common antimicrobials and a valuable scientific discovery in the twentieth century, caused a massive shift in pharmaceutical and veterinary sciences [6]. Molecules with antibiotic properties existed prior to humans producing manufactured antibiotics. However, widespread production of these compounds, along with synthetic derivatives of natural antibiotics, began in the twentieth century [7]. Soil actinobacteria and myxobacteria are the most significant fundamental reservoirs of active metabolites. More than 60% of natural antimicrobial compounds belong to actinobacteria and fungi [8].
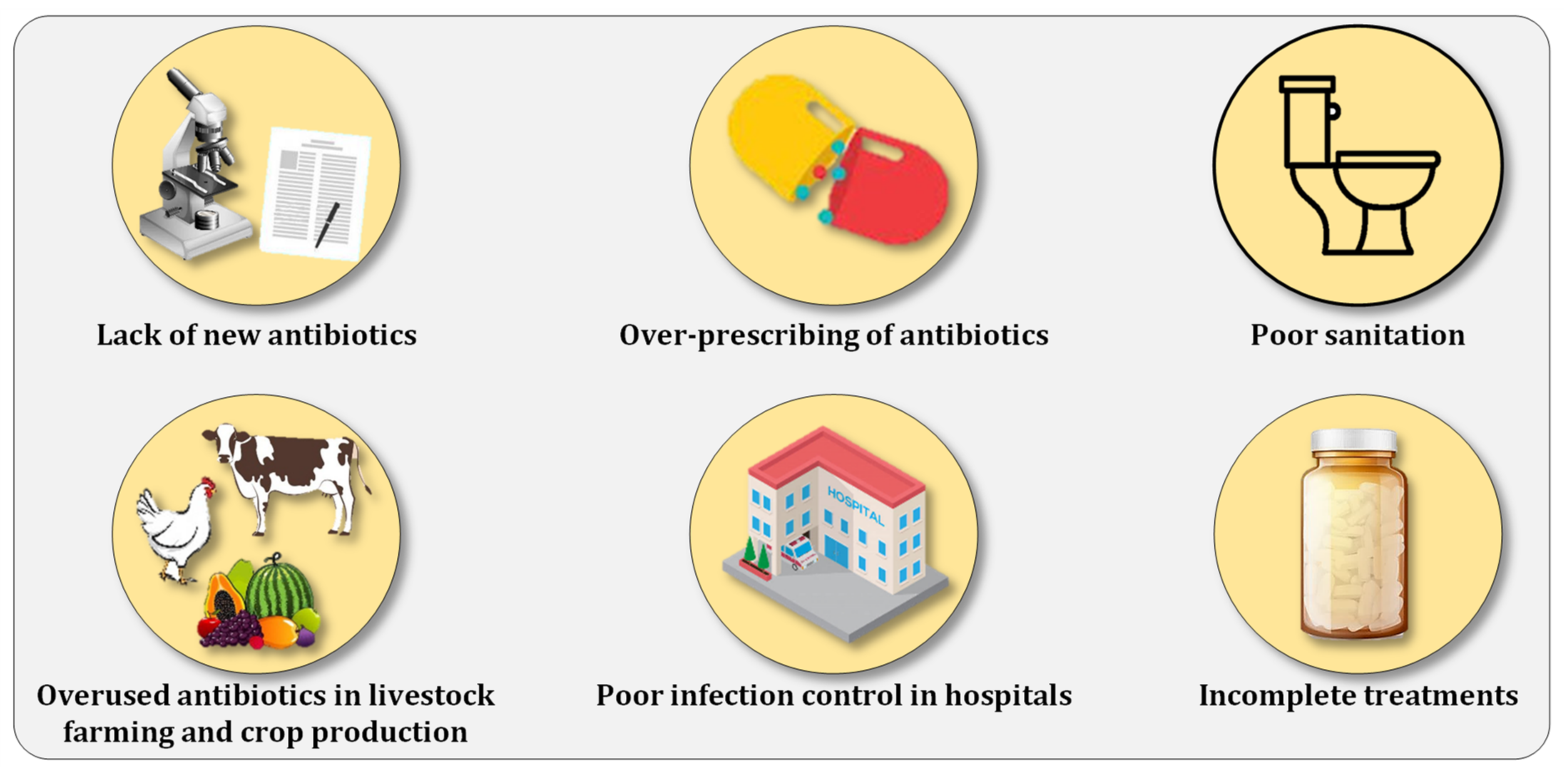
2. Combating Antibiotic Resistance Prevalence
References
- Nelkner, J.; Henke, C.; Lin, T.W.; Patzold, W.; Hassa, J.; Jaenicke, S.; Grosch, R.; Puhler, A.; Sczyrba, A.; Schluter, A. Effect of Long-Term Farming Practices on Agricultural Soil Microbiome Members Represented by Metagenomically Assembled Genomes (MAGs) and Their Predicted Plant-Beneficial Genes. Genes 2019, 10, 424.
- Nguyen, B.T.; Chen, Q.L.; He, J.Z.; Hu, H.W. Microbial regulation of natural antibiotic resistance: Understanding the protist-bacteria interactions for evolution of soil resistome. Sci. Total Environ. 2020, 705, 135882.
- Foo, J.L.; Ling, H.; Lee, Y.S.; Chang, M.W. Microbiome engineering: Current applications and its future. Biotechnol. J. 2017, 12, 1600099.
- Ochoa-Hueso, R. Global Change and the Soil Microbiome: A Human-Health Perspective. Front. Ecol. Evol. 2017, 5, 71.
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13.
- Bilal, M.; Mehmood, S.; Rasheed, T.; Iqbal, H.M.N. Antibiotics traces in the aquatic environment: Persistence and adverse environmental impact. Curr. Opin. Environ. Sci. Health 2020, 13, 68–74.
- Larsson, D.G. Antibiotics in the environment. Ups. J. Med. Sci. 2014, 119, 108–112.
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382.
- Baguer, A.J.; Jensen, J.; Krogh, P.H. Effects of the antibiotics oxytetracycline and tylosin on soil fauna. Chemosphere 2000, 40, 751–757.
- Isaacson, R.E.; Torrence, M.E. The Role of Antibiotics in Agriculture: This Report Is Based on a Colloquium Sponsored by the American Academy of Microbiology Held November 2–4, 2001, in Santa Fe, New Mexico; American Society for Microbiology: Washington, DC, USA, 2002.
- Callaway, T.R.; Lillehoj, H.; Chuanchuen, R.; Gay, C.G. Alternatives to Antibiotics: A Symposium on the Challenges and Solutions for Animal Health and Production. Antibiotics 2021, 10, 471.
- Akram, R.; Amin, A.; Hashmi, M.Z.; Wahid, A.; Mubeen, M.; Hammad, H.M.; Fahad, S.; Nasim, W. Fate of Antibiotics in Soil. In Antibiotics and Antibiotics Resistance Genes in Soils; Springer: Cham, Switzerland, 2017; pp. 207–220.
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759.
- Kumar, K.; Gupta, S.; Chander, Y.; Singh, A.K. Antibiotic Use in Agriculture and Its Impact on the Terrestrial Environment. Adv. Agron. 2005, 87, 1–54.
- Shen, Y.; Stedtfeld, R.D.; Guo, X.; Bhalsod, G.D.; Jeon, S.; Tiedje, J.M.; Li, H.; Zhang, W. Pharmaceutical exposure changed antibiotic resistance genes and bacterial communities in soil-surface- and overhead-irrigated greenhouse lettuce. Environ. Int. 2019, 131, 105031.
- Zalewska, M.; Blazejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 610656.
- CDC. About Antibiotic Resistance, 2021, Last Updated: 13 December 2021. Available online: https://www.cdc.gov/drugresistance/about.html (accessed on 2 January 2022).
- WHO. Antibiotic Resistance. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 31 July 2020).
- World Bank. Antimicrobial Resistance (AMR). 2021. Available online: https://www.worldbank.org/en/topic/health/brief/antimicrobial-resistance-amr (accessed on 13 May 2021).
- CDC. 2019 AR Threats Report. 2021. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 23 November 2021).
- WHO. Antimicrobial Resistance. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 17 November 2021).
- WHO. Infographics: Antibiotic Resistance. 2015. Available online: https://apps.who.int/mediacentre/events/2015/world-antibiotic-awareness-week/infographics/en/index.html (accessed on 22 November 2015).
- Wang, J.; Wang, L.; Zhu, L.; Wang, J.; Xing, B. Antibiotic resistance in agricultural soils: Source, fate, mechanism and attenuation strategy. Crit. Rev. Environ. Sci. Technol. 2020, 43, 1835438.
- Tyrrell, C.; Burgess, C.M.; Brennan, F.P.; Walsh, F. Antibiotic resistance in grass and soil. Biochem. Soc. Trans. 2019, 47, 477–486.
- Smalla, K.; Cook, K.; Djordjevic, S.P.; Klumper, U.; Gillings, M. Environmental dimensions of antibiotic resistance: Assessment of basic science gaps. FEMS Microbiol. Ecol. 2018, 94, fiy195.
- Jiao, S.; Chen, W.; Wei, G. Resilience and Assemblage of Soil Microbiome in Response to Chemical Contamination Combined with Plant Growth. Appl. Environ. Microbiol. 2019, 85, e02523-18.
- Zhang, Y.J.; Hu, H.W.; Yan, H.; Wang, J.T.; Lam, S.K.; Chen, Q.L.; Chen, D.; He, J.Z. Salinity as a predominant factor modulating the distribution patterns of antibiotic resistance genes in ocean and river beach soils. Sci. Total Environ. 2019, 668, 193–203.
- Hill, K.E.; Top, E.M. Gene transfer in soil systems using microcosms. FEMS Microbiol. Ecol. 1998, 25, 319–329.
- Beceiro, A.; Tomas, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230.
- Nayiga, S.; Kayendeke, M.; Nabirye, C.; Willis, L.D.; Chandler, C.I.R.; Staedke, S.G. Use of antibiotics to treat humans and animals in Uganda: A cross-sectional survey of households and farmers in rural, urban and peri-urban settings. JAC Antimicrob. Resist. 2020, 2, dlaa082.
- Madaras-Kelly, K. Optimizing antibiotic use in hospitals: The role of population-based antibiotic surveillance in limiting antibiotic resistance. Insights from the society of infectious diseases pharmacists. Pharmacotherapy 2003, 23, 1627–1633.
- Kromker, V.; Leimbach, S. Mastitis Treatment-Reduction in antibiotic usage in dairy cows. Reprod. Domest. Anim. 2017, 52, 21–29.
- Levy, S. Reduced antibiotic use in livestock: How Denmark tackled resistance. Environ. Health Perspect. 2014, 122, A160–A165.
- Mevius, D.; Heederik, D. Reduction of antibiotic use in animals “let’s go Dutch”. J. Verbrauch. Lebensm. 2014, 9, 177–181.
- Sneeringer, S.; Short, G.; MacLachlan, M.; Bowman, M. Impacts on Livestock Producers and Veterinarians of FDA Policies on Use of Medically Important Antibiotics in Food Animal Production. Appl. Econ. Perspect. Policy 2020, 42, 674–694.
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7, 1881.
- Lhermie, G.; Grohn, Y.T.; Raboisson, D. Addressing Antimicrobial Resistance: An Overview of Priority Actions to Prevent Suboptimal Antimicrobial Use in Food-Animal Production. Front. Microbiol. 2016, 7, 2114.
- Samanta, I.; Bandyopadhyay, S. The emergence of antimicrobial-resistant bacteria in livestock, poultry and agriculture. In Antimicrobial Resistance in Agriculture; Academic Press: Cambridge, MA, USA, 2020; p. 1927.
- Mshana, S.E.; Sindato, C.; Matee, M.I.; Mboera, L.E.G. Antimicrobial Use and Resistance in Agriculture and Food Production Systems in Africa: A Systematic Review. Antibiotics 2021, 10, 976.
- Robert, P.C. Precision agriculture: A challenge for crop nutrition management. Plant Soil 2002, 247, 143–149.
- Bongiovanni, R.; Lowenberg-Deboer, J. Precision Agriculture and Sustainability. Precis. Agric. 2004, 5, 359–387.
- Zarei, M.; Kaviani Rad, A. Covid-19, Challenges and Recommendations in Agriculture. J. Bot. Res. 2020, 2, 1841.
- Du, L.; Liu, W. Occurrence, fate, and ecotoxicity of antibiotics in agro-ecosystems. A review. Agron. Sustain. Dev. 2011, 32, 309–327.
- Sodhi, K.K.; Kumar, M.; Singh, D.K. Insight into the amoxicillin resistance, ecotoxicity, and remediation strategies. J. Water Process. Eng. 2021, 39, 101858.
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 1–11.
- Gu, S.; Kang, X.; Wang, L.; Lichtfouse, E.; Wang, C. Clay mineral adsorbents for heavy metal removal from wastewater: A review. Environ. Chem. Lett. 2018, 17, 629–654.
- Han, H.; Rafiq, M.K.; Zhou, T.; Xu, R.; Masek, O.; Li, X. A critical review of clay-based composites with enhanced adsorption performance for metal and organic pollutants. J. Hazard. Mater. 2019, 369, 780–796.
- Adeyemo, A.A.; Adeoye, I.O.; Bello, O.S. Adsorption of dyes using different types of clay: A review. Appl. Water Sci. 2015, 7, 543–568.
- Mustapha, S.; Ndamitso, M.M.; Abdulkareem, A.S.; Tijani, J.O.; Mohammed, A.K.; Shuaib, D.T. Potential of using kaolin as a natural adsorbent for the removal of pollutants from tannery wastewater. Heliyon 2019, 5, e02923.
- Yang, W.; Wu, Y.; Zhang, L.; Jiang, J.; Feng, L. Removal of five selected pharmaceuticals by coagulation in the presence of dissolved humic acids and kaolin. Desalination Water Treat. 2014, 54, 1134–1140.
- Ugwu, I.M.; Igbokwe, O.A. Sorption of heavy metals on clay minerals and oxides: A review. Adv. Sorpt. Process Appl. 2019, 23, 80989.
- Yan, W.; Bai, R.; Wang, S.; Tian, X.; Li, Y.; Wang, S.; Yang, F.; Xiao, Y.; Lu, X.; Zhao, F. Antibiotic resistance genes are increased by combined exposure to sulfamethoxazole and naproxen but relieved by low-salinity. Environ. Int. 2020, 139, 105742.
- Liu, M.; Li, Q.; Sun, H.; Jia, S.; He, X.; Li, M.; Zhang, X.-X.; Ye, L. Impact of salinity on antibiotic resistance genes in wastewater treatment bioreactors. Chem. Eng. J. 2018, 338, 557–563.
- Al-Mashhadany, D.A. Detection of antibiotic residues among raw beef in Erbil City (Iraq) and impact of temperature on antibiotic remains. Ital. J. Food Saf. 2019, 8, 7897.
- Zhang, Y.; Liu, H.; Dai, X.; Cai, C.; Wang, J.; Wang, M.; Shen, Y.; Wang, P. Impact of application of heat-activated persulfate oxidation treated erythromycin fermentation residue as a soil amendment: Soil chemical properties and antibiotic resistance. Sci. Total Environ. 2020, 736, 139668.
- Xu, R.; Zhang, Y.; Xiong, W.; Sun, W.; Fan, Q.; Zhaohui, Y. Metagenomic approach reveals the fate of antibiotic resistance genes in a temperature-raising anaerobic digester treating municipal sewage sludge. J. Clean. Prod. 2020, 277, 123504.
- Sun, C.; Li, W.; Chen, Z.; Qin, W.; Wen, X. Responses of antibiotics, antibiotic resistance genes, and mobile genetic elements in sewage sludge to thermal hydrolysis pre-treatment and various anaerobic digestion conditions. Environ. Int. 2019, 133, 105156.
- Liao, H.; Lu, X.; Rensing, C.; Friman, V.P.; Geisen, S.; Chen, Z.; Yu, Z.; Wei, Z.; Zhou, S.; Zhu, Y. Hyperthermophilic Composting Accelerates the Removal of Antibiotic Resistance Genes and Mobile Genetic Elements in Sewage Sludge. Environ. Sci. Technol. 2018, 52, 266–276.
- Deng, W.; Zhang, A.; Chen, S.; He, X.; Jin, L.; Yu, X.; Yang, S.; Li, B.; Fan, L.; Ji, L.; et al. Heavy metals, antibiotics and nutrients affect the bacterial community and resistance genes in chicken manure composting and fertilized soil. J. Environ. Manag. 2020, 257, 109980.
- Gou, M.; Hu, H.W.; Zhang, Y.J.; Wang, J.T.; Hayden, H.; Tang, Y.Q.; He, J.Z. Aerobic composting reduces antibiotic resistance genes in cattle manure and the resistome dissemination in agricultural soils. Sci. Total Environ. 2018, 612, 1300–1310.
- Youngquist, C.P.; Mitchell, S.M.; Cogger, C.G. Fate of Antibiotics and Antibiotic Resistance during Digestion and Composting: A Review. J. Environ. Qual. 2016, 45, 537–545.
- Keenum, I.; Williams, R.K.; Ray, P.; Garner, E.D.; Knowlton, K.F.; Pruden, A. Combined effects of composting and antibiotic administration on cattle manure-borne antibiotic resistance genes. Microbiome 2021, 9, 81.
- Sardar, M.F.; Zhu, C.; Geng, B.; Ahmad, H.R.; Song, T.; Li, H. The fate of antibiotic resistance genes in cow manure composting: Shaped by temperature-controlled composting stages. Bioresour. Technol. 2021, 320, 124403.
- Gao, M.; Qiu, T.; Sun, Y.; Wang, X. The abundance and diversity of antibiotic resistance genes in the atmospheric environment of composting plants. Environ. Int. 2018, 116, 229–238.
- Guerin, T.F. The differential removal of aged polycyclic aromatic hydrocarbons from soil during bioremediation. Environ. Sci. Pollut. Res. 2000, 7, 19–26.
- Imam, A.; Suman, S.K.; Ghosh, D.; Kanaujia, P.K. Analytical approaches used in monitoring the bioremediation of hydrocarbons in petroleum-contaminated soil and sludge. TrAC Trends Anal. Chem. 2019, 118, 50–64.
- Stroud, J.L.; Paton, G.I.; Semple, K.T. Microbe-aliphatic hydrocarbon interactions in soil: Implications for biodegradation and bioremediation. J. Appl. Microbiol. 2007, 102, 1239–1253.
- Diplock, E.E.; Mardlin, D.P.; Killham, K.S.; Paton, G.I. Predicting bioremediation of hydrocarbons: Laboratory to field scale. Environ. Pollut. 2009, 157, 1831–1840.
- Ghazali, F.M.; Rahman, R.N.Z.A.; Salleh, A.B.; Basri, M. Biodegradation of hydrocarbons in soil by microbial consortium. Int. Biodeterior. Biodegrad. 2004, 54, 61–67.
- Gargouri, B.; Karray, F.; Mhiri, N.; Aloui, F.; Sayadi, S. Bioremediation of petroleum hydrocarbons-contaminated soil by bacterial consortium isolated from an industrial wastewater treatment plant. J. Chem. Technol. Biotechnol. 2014, 89, 978–987.
- Guarino, C.; Spada, V.; Sciarrillo, R. Assessment of three approaches of bioremediation (Natural Attenuation, Landfarming and Bioagumentation—Assistited Landfarming) for a petroleum hydrocarbons contaminated soil. Chemosphere 2017, 170, 10–16.
- Muyzer, G.; Stams, A.J. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454.
- Castro, H.F.; Williams, N.H.; Ogram, A. Phylogeny of sulfate-reducing bacteria1. FEMS Microbiol. Ecol. 2000, 31, 1–9.
- Xu, Y.N.; Chen, Y. Advances in heavy metal removal by sulfate-reducing bacteria. Water Sci. Technol. 2020, 81, 1797–1827.
- Jong, T.; Parry, D.L. Removal of sulfate and heavy metals by sulfate reducing bacteria in short-term bench scale upflow anaerobic packed bed reactor runs. Water Res. 2003, 37, 3379–3389.
- Zhao, Q.; Li, X.; Xiao, S.; Peng, W.; Fan, W. Integrated remediation of sulfate reducing bacteria and nano zero valent iron on cadmium contaminated sediments. J. Hazard. Mater. 2021, 406, 124680.
- Barton, L.L.; Fauque, G.D. Biochemistry, Physiology and Biotechnology of Sulfate-Reducing Bacteria. Adv. Appl. Microbiol. 2009, 68, 41–98.
- Fang, H.; Oberoi, A.S.; He, Z.; Khanal, S.K.; Lu, H. Ciprofloxacin-degrading Paraclostridium sp. isolated from sulfate-reducing bacteria-enriched sludge: Optimization and mechanism. Water Res. 2021, 191, 116808.
- Zhang, H.; Song, S.; Jia, Y.; Wu, D.; Lu, H. Stress-responses of activated sludge and anaerobic sulfate-reducing bacteria sludge under long-term ciprofloxacin exposure. Water Res. 2019, 164, 114964.
- Zhang, H.; Jia, Y.; Khanal, S.K.; Lu, H.; Fang, H.; Zhao, Q. Understanding the Role of Extracellular Polymeric Substances on Ciprofloxacin Adsorption in Aerobic Sludge, Anaerobic Sludge, and Sulfate-Reducing Bacteria Sludge Systems. Environ. Sci. Technol. 2018, 52, 6476–6486.
- Jia, Y.; Khanal, S.K.; Shu, H.; Zhang, H.; Chen, G.H.; Lu, H. Ciprofloxacin degradation in anaerobic sulfate-reducing bacteria (SRB) sludge system: Mechanism and pathways. Water Res. 2018, 136, 64–74.
- Zhang, X.; Zhu, R.; Li, W.; Ma, J.; Lin, H. Genomic insights into the antibiotic resistance pattern of the tetracycline-degrading bacterium, Arthrobacter nicotianae OTC-16. Sci. Rep. 2021, 11, 15638.
- Maki, T.; Hasegawa, H.; Kitami, H.; Fumoto, K.; Munekage, Y.; Ueda, K. Bacterial degradation of antibiotic residues in marine fish farm sediments of Uranouchi Bay and phylogenetic analysis of antibiotic-degrading bacteria using 16S rDNA sequences. Fish. Sci. 2006, 72, 811–820.
- Hirth, N.; Topp, E.; Dörfler, U.; Stupperich, E.; Munch, J.C.; Schroll, R. An effective bioremediation approach for enhanced microbial degradation of the veterinary antibiotic sulfamethazine in an agricultural soil. Chem. Biol. Technol. Agric. 2016, 3, 1–11.
- Mojiri, A.; Baharlooeian, M.; Zahed, M.A. The Potential of Chaetoceros muelleri in Bioremediation of Antibiotics: Performance and Optimization. Int. J. Environ. Res. Public Health 2021, 18, 977.
- Wu, Y.; Feng, P.; Li, R.; Chen, X.; Li, X.; Sumpradit, T.; Liu, P. Progress in microbial remediation of antibiotic-residue contaminated environment. Sheng Wu Gong Cheng Xue Bao 2019, 35, 2133–2150.
- Zhi, D.; Yang, D.; Zheng, Y.; Yang, Y.; He, Y.; Luo, L.; Zhou, Y. Current progress in the adsorption, transport and biodegradation of antibiotics in soil. J. Environ. Manage. 2019, 251, 109598.
- Xue, W.; Zhou, Q.; Li, F. Bacterial community changes and antibiotic resistance gene quantification in microbial electrolysis cells during long-term sulfamethoxazole treatment. Bioresour. Technol. 2019, 294, 122170.
- Hua, T.; Li, S.; Li, F.; Ondon, B.S.; Liu, Y.; Wang, H. Degradation performance and microbial community analysis of microbial electrolysis cells for erythromycin wastewater treatment. Biochem. Eng. J. 2019, 146, 1–9.
- Zhang, X.; Li, R. Variation and distribution of antibiotic resistance genes and their potential hosts in microbial electrolysis cells treating sewage sludge. Bioresour. Technol. 2020, 315, 123838.
- Zhang, X.; Li, R. Electrodes bioaugmentation promotes the removal of antibiotics from concentrated sludge in microbial electrolysis cells. Sci. Total Environ. 2020, 715, 136997.
- Yan, W.; Xiao, Y.; Yan, W.; Ding, R.; Wang, S.; Zhao, F. The effect of bioelectrochemical systems on antibiotics removal and antibiotic resistance genes: A review. Chem. Eng. J. 2019, 358, 1421–1437.
- Dong, X.; Rao, D.; Tian, L.; Wang, Q.; Yang, K. A slurry microcosm study on the interaction between antibiotics and soil bacterial community. Heliyon 2020, 6, e03348.
- Naqvi, S.A.R.; Nadeem, S.; Komal, S.; Naqvi, S.A.A.; Mubarik, M.S.; Qureshi, S.Y.; Ahmad, S.; Abbas, A.; Zahid, M.; Raza, S.S. Antioxidants: Natural Antibiotics. In Antioxidants; IntechOpen: London, UK, 2019.
- Abd El-Ghany, W.A. Paraprobiotics and postbiotics: Contemporary and promising natural antibiotics alternatives and their applications in the poultry field. Open Vet. J. 2020, 10, 323–330.
- Gupta, R.; Sharma, S. Herbal antibiotics: A Review. Bull. Env. Pharmacol. Life Sci. 2020, 9, 136–142.
- Ionescu, M.I. Are Herbal Products an Alternative to Antibiotics. In Bacterial Pathogenesis and Antibacterial Control; IntechOpen: London, UK, 2018.
- Fit, N.; Gheorghe, R.; Rapuntean, S.; Flore, C.; Nadas, G. Antibacterial Effect of Essential Vegetal Extracts on Staphylococcus aureus Compared to Antibiotics. Not. Bot. Horti Agrobot. Cluj Napoca 2009, 37, 117–123.
- Awan, U.; Andleeb, D.S.; Kiyani, A.; Zafar, A.; Shafique, I.; Riaz, N.; Azhar, M.T.; Uddin, H. Antibacterial screening of traditional herbal plants and standard antibiotics against some human bacterial pathogens. Pak. J. Pharm. Sci. 2013, 26, 1109–1116.
- Saquib, S.A.; AlQahtani, N.A.; Ahmad, I.; Kader, M.A.; Al Shahrani, S.S.; Asiri, E.A. Evaluation and Comparison of Antibacterial Efficacy of Herbal Extracts in Combination with Antibiotics on Periodontal pathobionts: An in vitro Microbiological Study. Antibiotics 2019, 8, 89.
- Nweze, E.I.; Eze, E.E. Justification for the use of Ocimum gratissimum L in herbal medicine and its interaction with disc antibiotics. BMC Complement. Altern. Med. 2009, 9, 37.
- Sharifi-Rad, J. Herbal Antibiotics: Moving back into the mainstream as an alternative for “Superbugs”. Cell Mol. Biol. 2016, 62, 1–2.
- Martin, K.W.; Ernst, E. Herbal medicines for treatment of bacterial infections: A review of controlled clinical trials. Antimicrob. Chemother. 2003, 51, 241–246.
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395.
- Koczulla, R.; Bals, R. Antimicrobial Peptides: Current Status and Therapeutic Potential. Drugs 2003, 63, 389–406.
- Reddy, K.V.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547.
- Sang, Y.; Blecha, F. Antimicrobial peptides and bacteriocins: Alternatives to traditional antibiotics. Anim. Health Res. Rev. 2008, 9, 227–235.
- Leontiadou, H.; Mark, A.E.; Marrink, S.J. Antimicrobial Peptides in Action. J. Am. Chem. Soc. 2006, 128, 12156–12161.
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51.
- Malmsten, M. Antimicrobial peptides. Ups. J. Med. Sci. 2014, 119, 199–204.
- Wang, G.; Mishra, B.; Lau, K.; Lushnikova, T.; Golla, R.; Wang, X. Antimicrobial peptides in 2014. Pharmaceuticals 2015, 8, 123–150.
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575.
- Kosciuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzalkowska, N.; Jozwik, A.; Horbanczuk, J.; Krzyzewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970.
- Tamhankar, A.J.; Stalsby Lundborg, C. Antimicrobials and Antimicrobial Resistance in the Environment and Its Remediation: A Global One Health Perspective. Int. J. Environ. Res. Public Health 2019, 16, 4614.
- Rad, A.; Zarei, M.; Pourghasemi, H.; Tiefenbacher, J. The COVID-19 crisis and its consequences for global warming and climate change. In Computers in Earth and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2022; pp. 377–385.
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795.




