Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Workie Anley Zegeye | -- | 1108 | 2022-04-25 08:48:15 | | | |
| 2 | Camila Xu | Meta information modification | 1108 | 2022-04-25 08:56:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zegeye, W.; , .; Zhang, Y.; Liyong, C. CRISPR/Cas9 System. Encyclopedia. Available online: https://encyclopedia.pub/entry/22223 (accessed on 02 March 2026).
Zegeye W, , Zhang Y, Liyong C. CRISPR/Cas9 System. Encyclopedia. Available at: https://encyclopedia.pub/entry/22223. Accessed March 02, 2026.
Zegeye, Workie, , Yingxin Zhang, Cao Liyong. "CRISPR/Cas9 System" Encyclopedia, https://encyclopedia.pub/entry/22223 (accessed March 02, 2026).
Zegeye, W., , ., Zhang, Y., & Liyong, C. (2022, April 25). CRISPR/Cas9 System. In Encyclopedia. https://encyclopedia.pub/entry/22223
Zegeye, Workie, et al. "CRISPR/Cas9 System." Encyclopedia. Web. 25 April, 2022.
Copy Citation
CRISPR/Cas9 is an endonuclease of DNA that splits the invading phage DNA into pieces and then incorporates it into the CRISPR set as a spacer. It was implemented efficiently in plants in 2013, and in five original research articles, the CRISPR/Cas9 scheme in rice was efficiently recorded.
CRISPR/Cas9
Cas variants
genome editing
1. CRISPR/Cas9 System and Its Components
Initially, CRISPR/Cas9 was discovered as an adaptive immune system of archaea and bacteria. It was first reported by [1], and researchers discovered a means of exploiting it as a gene-editing technology. This technique can identify a particular site in a target gene in a highly efficient, unique, and flexible manner [2]. In roughly 50% of bacteria and 90% of archaea, CRISPR/Cas9 active immune mechanisms exist [3]. CRISPR/Cas9 is an endonuclease of DNA that splits the invading phage DNA into pieces and then incorporates it into the CRISPR set as a spacer. It was implemented efficiently in plants in 2013, and in five original research articles, the CRISPR/Cas9 scheme in rice was efficiently recorded [4][5][6][7][8]. Consequently, CRISPR/Cas9 technologies have proved to be an essential genome-editing method for rice.
Nowadays, the editing system of CRISPR/Cas9 is the most common mechanism in plant biology for the genome editing process [9][10] because of its uncomplicatedness, flexibility, and efficiency. Two CRISPR/Cas9 components: the Cas9 protein and a short RNA molecule (sgRNA), together form a ribonucleoprotein complex (Figure 1). The sgRNA is made up of 18 to 21 nucleotides that are designed to target specific sites in the genome (protospacer). A G-rich (5′-NGG-3′) protospacer-adjacent motif (PAM) should be found downstream of the target site [1]. tracrRNA (trans-activating CRISPR RNA) and crRNA (CRISPR-derived RNA) are the two principal components of guide RNA. In essence, crRNA has homology area that enables it to integrate with tracrRNA. The tracrRNA has a stem-loop shape associated with the Cas9 protein. The crRNA and tracrRNA should be designed as sgRNA in the CRISPR/Cas9 gene-editing scheme to direct Cas9 dsDNA cleavage at the targeted site [1]. Following the appropriate site identification by sgRNA based on the Watson–Crick base-pairing rule, the Cas9-sgRNA complex moves through the genome and produces a double-stranded break (DSB) [3]. During cleavage site repairing, the error-prone non-homologous end-joining (NHEJ) pathway regularly leaves a lesion in the form of a minor InDel or substitution upstream of PAM. Such mutations can induce frame-shift mutations in the coding sequence of the gene and cause a premature stop codon, resulting in a loss or gain of function in mutants. This achievement has unlocked numerous other possibilities for scientists to obtain more knowledge on plant biological systems [11].
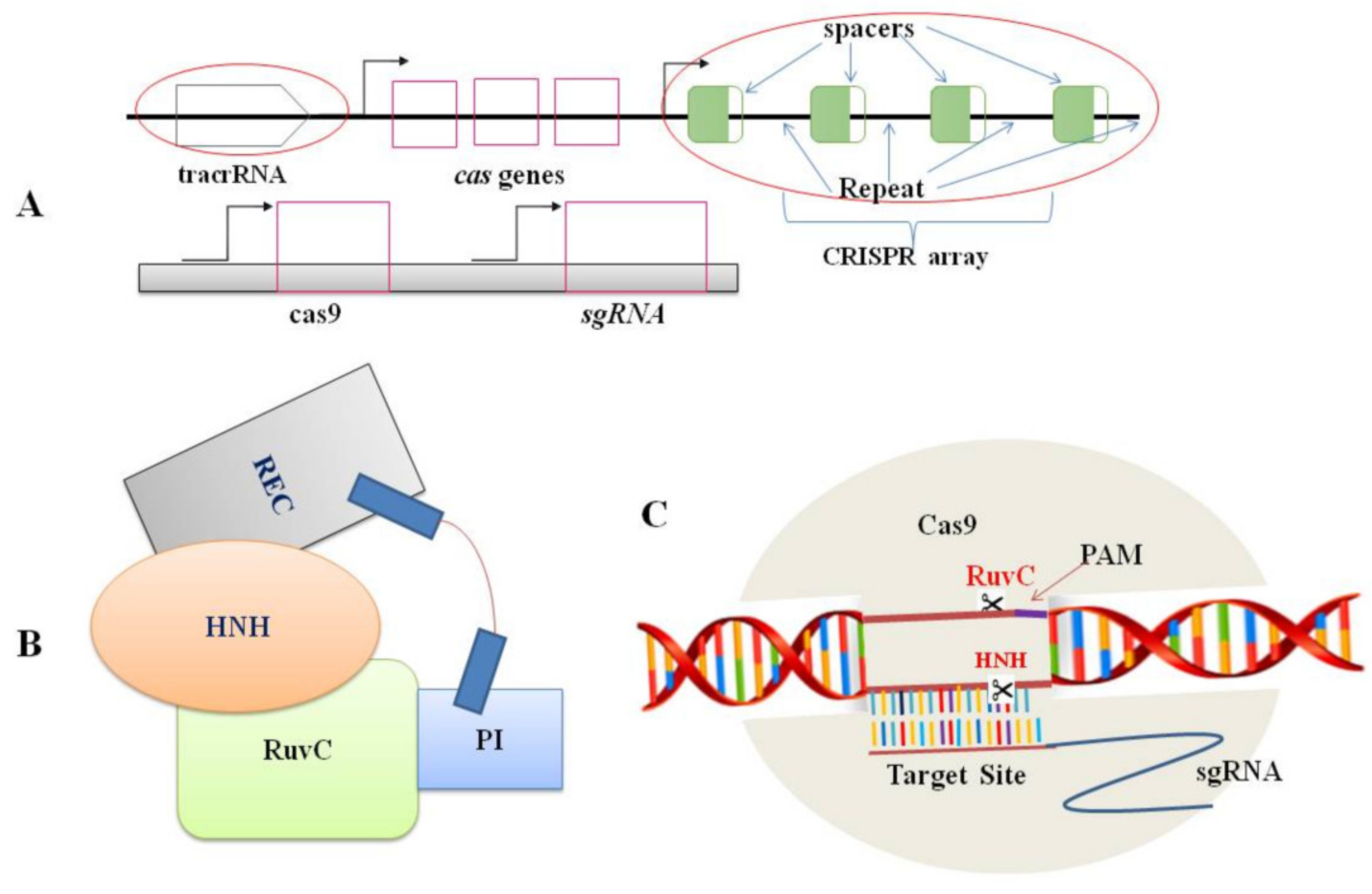 Figure 1. Components of CRISPR/Cas9 system: (A). Genomic structures of the CRISPR/Cas system (top) and the engineered CRISPR/Cas9 system (bottom); (B). A schematic representation of the Cas9 protein structure. Domains includes REC (large recognition lobe) and RuvC (a nuclease domain), which is linked with an arginine-rich region. HNH is a second nuclease domain. PI is PAM-interacting domain; (C). The conformation of the Cas9–sgRNA complex in the process of DNA cleavage. The Cas9 endonuclease is targeted to DNA by a guide RNA which can be supplied as a two-part system consisting of crRNA and tracrRNA or as a single guide RNA, where the crRNA and tracrRNA are connected by a linker. Target recognition is facilitated by the protospacer-adjacent motif (PAM). Cleavage occurs on both strands (scissors) 3 bp upstream of the PAM.
Figure 1. Components of CRISPR/Cas9 system: (A). Genomic structures of the CRISPR/Cas system (top) and the engineered CRISPR/Cas9 system (bottom); (B). A schematic representation of the Cas9 protein structure. Domains includes REC (large recognition lobe) and RuvC (a nuclease domain), which is linked with an arginine-rich region. HNH is a second nuclease domain. PI is PAM-interacting domain; (C). The conformation of the Cas9–sgRNA complex in the process of DNA cleavage. The Cas9 endonuclease is targeted to DNA by a guide RNA which can be supplied as a two-part system consisting of crRNA and tracrRNA or as a single guide RNA, where the crRNA and tracrRNA are connected by a linker. Target recognition is facilitated by the protospacer-adjacent motif (PAM). Cleavage occurs on both strands (scissors) 3 bp upstream of the PAM.2. Mechanism of the CRISPR/Cas9 System
In three steps, namely adaptation/acquisition, expression, and interference, CRISPR/Cas9 identifies and targets the genetic material of foreign DNA [12][13][14][15]. The adaptation/acquisition phase includes the recognition, invasion, and binding of donor DNA that are cut into small segments and combined within the CRISPR locus. Then, the CRISPR locus is transcribed to create crRNA that directs the intended effect or endonucleases to attack viral items by complementary base pairing [16][17]. Since the protospacer contains a G-rich base pair (5′-NGG-3′), PAMs are used as recognition motifs for the adaptation/acquisition of the targeted site. During the second step of the CRISPR/Cas9 execution mechanism, the lengthy Pre-crRNA is deliberately transcribed from the CRISPR nucleus and reproduced into crRNAs using Cas9 proteins. Recently, researchers revealed that tracrRNA is also involved in Streptococcus pyogenes pre-crRNA processing [18]. The tracrRNA is associated with the repeat crRNA site through complementary base pairing and allows precrRNA to be processed in crRNA [19]. The processed crRNAs join the associated antiviral defense complex of CRISPR and help to identify and parse a particular target area of donor DNA [19]. At the final steps, i.e., interference, the system needs a Cas9 protein [20][21] so that the sgRNA can guide the cleavage of the Cas9 protein complex from the particular target area; this creates immunity from pathogen attacks [12][13].
3. The Advantages and Drawbacks of the CRISPR/Cas9 System
Until recently, the CRISPR/Cas9 tool was thought to be the best option for genome editing (GE) in plants, but it still has some drawbacks that limit its widespread application (Figure 2).
 Figure 2. The advantages and disadvantages of the CRISPR/Cas9 system over other approaches for genome editing. (A). Conventional gene targeting. (B). ZNFs and TALENs. (C). CRISPR/SpCas9. (D). CRISPR/NmCas9. The red arrow indicates the corresponding gene editing method with its features, advantages, and disadvantages.
Figure 2. The advantages and disadvantages of the CRISPR/Cas9 system over other approaches for genome editing. (A). Conventional gene targeting. (B). ZNFs and TALENs. (C). CRISPR/SpCas9. (D). CRISPR/NmCas9. The red arrow indicates the corresponding gene editing method with its features, advantages, and disadvantages.The following are a list of the major issues with CRISPR/Cas9 and the advantages of CRISPR/Cas variants:
-
The CRISPR/Cas9 system’s large size limits its editing efficiency, and it is not suitable for packing into viral vectors for delivery to somatic tissues. For efficient plant GE, a smaller-sized CRISPR/Cas is required;
-
SpCas9 involves a 5′-NGG-3′ PAM next to a 20 nt DNA target site where it only distinguishes the NGG PAM sequence, which limits its effectiveness when compared to new CRISPR/Cas variants. NG-Cas9 is more active and there is a newer variant, SpRYCas9 that is almost PAM-less. The broad PAM compatibility of SpRY greatly expands the targeting scope of CRISPR-based tools in plant genome engineering;
-
CRISPR/Cas9 has the potential to incorporate a large number of off-target mutations into the genome. However, by identifying various PAMs, new CRISPR/Cas variants have achieved better editing efficiency (fewer off-target mutations) of target bases in the sequence of interest;
-
CRISPR/Cas9 generates mutations at non-specific loci that are homologous to target sites;
-
CRISPR/Cas9-made mutant plants via Agrobacterium-mediated transformation systems are more expensive, time consuming, and resource intensive. The use of tissue culture-free genome editing systems, on the other hand, has the potential to improve efficiency;
-
The commercialization of transgenic crops expressing CRISPR/Cas9 faces challenges in a number of countries, owing primarily to development costs and constraints imposed by regulatory systems for the field release of genetically modified organisms.
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821.
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997.
- Makarova, K.S.; Wolf, Y.; Alkhnbashi, O.S.; Costa, F.; Shah, S.; Saunders, S.; Barrangou, R.; Brouns, S.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736.
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688.
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.-L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient Genome Editing in Plants Using a CRISPR/Cas System. Cell Res. 2013, 23, 1229–1232.
- Jiang, W.Z.; Zhou, H.B.; Bi, H.H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, Tobacco, Sorghum, and rice. Nucleic Acids Res. 2013, 41, e188.
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236.
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 2013, 6, 1975–1983.
- Belhaj, K.; Chaparro Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84.
- Weeks, D.P.; Spalding, M.H.; Yang, B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 2016, 14, 483–495.
- Ricroch, A.; Clairand, P.; Harwood, W. Use of CRISPR systems in plant genome editing: Toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182.
- Garneau, J.; Dupuis, M.; Villion, M.; Romero, D.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71.
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010, 11, 181–190.
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170.
- Van der Oost, J.; Jore, M.M.; Westra, E.R.; Lundgren, M.; Brouns, S.J. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009, 34, 401–407.
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712.
- Yosef, I.; Goren, M.G.; Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012, 40, 5569–5576.
- Karvelis, T.; Gasiunas, G.; Miksys, A.; Barrangou, R.; Horvath, P.; Siksnys, V. crRNA and tracrRNA guide Cas9-mediated DNA interference in streptococcus thermophilus. RNA Biol. 2013, 10, 841–851.
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, I.J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607.
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956.
- Zetsche, B.; Gootenberg, J.; Abudayyeh, O.; Slaymaker, J.; Makarova, K.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of Class 2 CRISPR-Cas system. Cell 2015, 163, 759–771.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.4K
Revisions:
2 times
(View History)
Update Date:
25 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





