Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wei, L.; Nhieu, J.; Lin, Y. CRABP1. Encyclopedia. Available online: https://encyclopedia.pub/entry/22022 (accessed on 07 February 2026).
Wei L, Nhieu J, Lin Y. CRABP1. Encyclopedia. Available at: https://encyclopedia.pub/entry/22022. Accessed February 07, 2026.
Wei, Li-Na, Jennifer Nhieu, Yu-Lung Lin. "CRABP1" Encyclopedia, https://encyclopedia.pub/entry/22022 (accessed February 07, 2026).
Wei, L., Nhieu, J., & Lin, Y. (2022, April 20). CRABP1. In Encyclopedia. https://encyclopedia.pub/entry/22022
Wei, Li-Na, et al. "CRABP1." Encyclopedia. Web. 20 April, 2022.
Copy Citation
The emerging role of Cellular Retinoic Acid Binding Protein 1 (CRABP1) as a mediator of non-canonical activities of retinoic acid (RA) and relevance to human diseases.
CRABP1
retinoic acid
neurodegeneration
inflammation
1. Introduction: Canonical and Non-Canonical Activities of All-Trans Retinoic Acid (atRA)
Vitamin A (also known as retinol) is an essential nutrient required for almost all physiological processes [1]. The profound effects of vitamin A are elicited mainly through atRA, as well as its various isomers. Through decades of studies, it has been established that atRA, as the principal active metabolite of vitamin A, executes its activities through binding to its nuclear receptors, RA receptors (RARs), which usually pair with Retinoid X Receptors (RXRs), can bind the cis isomers of atRA. These RAR/RXR pairs, in various combinations, act to regulate the transcription of numerous target genes that harbor RA response elements (RAREs) in their regulatory regions [2][3]. RAR/RXR pairs often also act together with other transcription factors to confer further specificity in the expression of target genes, resulting in the tight regulation of specific cellular processes such as proliferation [4], differentiation [5], apoptosis [6], and other physiological functions. These ultimately ensure the homeostasis of most organ systems/physiological processes [2][7]. Dysregulated RA signaling often leads to disease conditions [8][9][10]. These RAR-mediated activities of RA, which occur in the nucleus to regulate the execution of genetic programs, generally span an extended period of time (days to years) and are referred to as canonical activities of RA [11]. An extensive body of work has determined that the cellular retinoid-binding proteins (CRABPs) I and II facilitate these canonical activities of RA. Rigorous biochemical studies have characterized classical CRABP1 functions in RA binding, sequestration, and metabolism via cytochrome (CYP) P-450 enzymes [12][13], (reviewed in-depth in [14][15][16][17]) while CRABP2 is responsible for the transport of RA to the nucleus [14][15][18][19].
In 2008, a study first reported a novel activity (effect) of atRA that occurred rapidly (within minutes) to alter the protein phosphorylation status of a transcription factor TR2 in maintaining stem cell proliferation and stemness potential [20]. Subsequent studies [21][22][23] further documented similar activities of atRA that shared several features: (1) RAR-independence, (2) occurring in the cytosol without altering gene expression, and (3) rapid (typically within minutes) action. These novel activities of atRA were collectedly referred to as “non-canonical” and were later found to be mediated by the Cellular Retinoic Acid Binding Protein 1 (CRABP1) [22]. These CRABP1-mediated non-canonical activities of atRA were ultimately validated in careful studies of Crabp1 knockout (CKO) mice and cultures, which also revealed the physiological/disease relevance of CRABP1 [24][25][26][27][28][29][30][31][32][33].
Extensive molecular and cell biological studies have identified specific cytosolic signaling pathways that can be targeted by CRABP1 in a cell context-dependent manner. It is believed that CRABP1 functions as a signal integrator by forming various specific RA-regulated signaling protein complexes (signalsomes) in different cells to modulate specific cellular processes/functions.
2. CRABP1-Signalsomes
CRABP1 is the most highly conserved retinoid-binding protein among all the known binding proteins and nuclear receptors for retinoids. CRABP1 binds, specifically, atRA with a high affinity (<1 nM) [34][35][36][37]. Given its high affinity toward atRA and cytosolic distribution, CRABP1 has been proposed and shown to sequester the poorly soluble RA from the aqueous cytosolic environment [12][13][14][15][16][17][30]. This led to the belief that CRABP1 would function to control RA availability in the cell, which indeed was supported by several molecular studies, by altering the expression level of CRABP1, that documented subsequent changes in the expression of RA-responding genes [38][39]. As introduced earlier, CRABP1 could participate in RA metabolism by delivering RA to CYP P-450 metabolic enzymes and microsomes via protein-protein interactions and substrate channeling [15][40]. However, the physiological role of CRABP1 in mediating the newly observed, non-canonical activity has remained largely elusive. Only recently, studies of CKO mice and cultures in various physiological/pathological conditions (see the following section) began to shed light on multiple functional roles of CRABP1 in modulating specific cellular processes, which contributed to the “non-canonical” activity. The fact that CRABP1 is important for multiple signaling pathways is consistent with the extremely high conservation of its amino acid sequence across animal species. Figure 1 shows the reported amino acid sequence alignment of CRABP1 among five animal species including human [41], pig [42], rat [43], mouse [44], and bovine [45]. Importantly, there is only a single residue, at position 86, that is not conserved, with alanine in human and pig sequences and proline in mouse, rat, and bovine sequences (Figure 1).

Figure 1. CRABP1 sequence alignment across mammals. Reported CRABP1 protein sequences from the Uniprot database of human (ID: P29762) [41], pig (ID: B3F0B7) [42], rat (ID: P62966) [43], mouse (ID: P62965) [44], and bovine (ID: P62964) [45] were aligned using the ClustalWS alignment algorithm in Jalview. Only the residue at position 86 (indicated by bold text and arrow ↓) is not conserved and exists either as an alanine (A) in human and pig sequences or as a proline (P) in bovine, rat, and mouse sequences.
The extreme conservation of CRABP1 during evolution would suggest important functional constraints. The evidence for this notion was obtained in careful studies of CKO mouse phenotypes (see later). Mechanistic details were provided in biochemical and cellular studies that first revealed specific context-dependent “binding partners” of CRABP1, which were rigorously defined according to at least two criteria: (a) specific and direct binding to CRABP1, which could be validated in vitro, and (b) forming specific cytosolic protein complexes that could be validated in vivo. Functional consequences of these CRABP1-containing protein complexes were each found to be capable of modulating certain specific cytosolic signaling pathways in a particular cell type. These CRABP1-containing protein complexes are therefore referred to as CRABP1-signalsomes. Currently, two types of CRABP1-signalsomes have been identified, which are discussed in the following sections.
2.1. CRABP1-MAPK (RAF-MEK-ERK) Signalsome in Stem Cells, Cancers
A specific Crabp1-signaling complex was first proposed after studying embryonal carcinoma (EC) and embryonic stem (ES) cells that were stimulated by a physiological concentration (10 nM) of atRA to modulate their proliferation/differentiation (reviewed in [46][47]). The initial study detected a very rapid (within minutes) response of these cells to atRA administration, which occurred in the cytosol and involved a mitogen-activated protein kinase (MAPK) pathway to modify target proteins for specific post-translational modifications [20][21][22][23]. This atRA-elicited signal was found to involve CRABP1, and could rapidly (within minutes) alter (dampen) the activity of the initiating kinase of the MAPK pathway, which is the rapidly Accelerated Fibrosarcoma (RAF) kinase and is a cell membrane-anchored kinase activated by the mitogenic signal Ras GTPase [48]. The MAPK kinase signaling cascade is comprised of Ras GTPase which activates RAF, then mitogen-activated protein kinase kinase (MEK), and then extracellular-signal-regulated kinase (ERK). Activation of this signaling pathway generally leads to cell proliferation and growth for stem/progenitor cells [48]. Through biochemical and molecular studies, it is now established that CRABP1 competes with Ras by directly interacting with RAF at its Ras-binding domain, thereby dampening MAPK signal propagation and ultimately modulating (reducing) cell proliferation of ES, EC, and neural stem cell (NSC) [22][25][29]. The proposed mechanistic model for CRABP1-MAPK signalsome is shown in Figure 2.
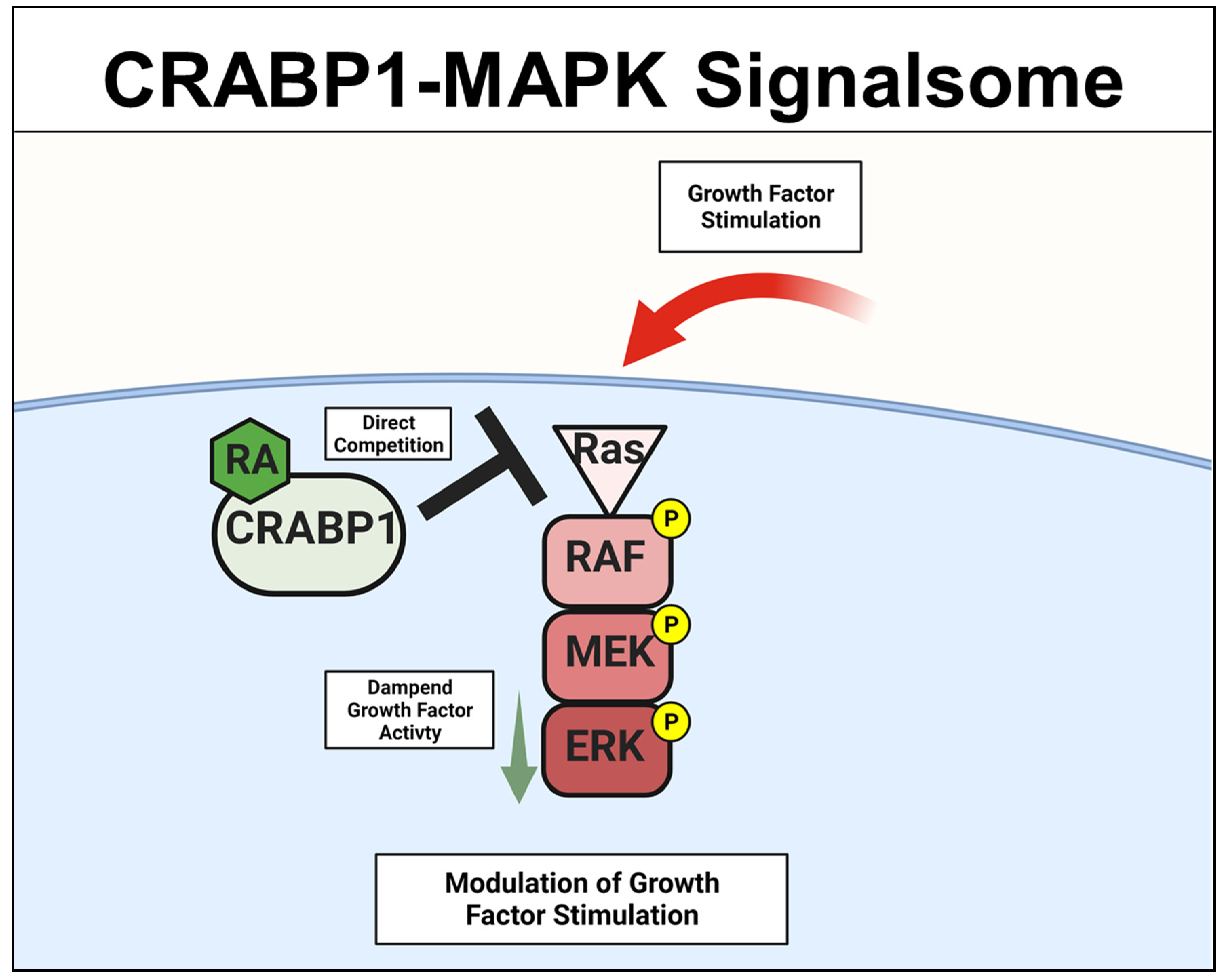
Figure 2. CRABP1-MAPK signalsome. The action of CRABP1-signalsome in growth-factor stimulated MAPK activity is mediated by its direct competition with Ras, resulting in dampened MAPK activation. CRABP1: Cellular Retinoic Acid Binding Protein 1, RA: retinoic acid, RAF: rapidly Accelerated Fibrosarcoma, MEK: mitogen-activated protein kinase kinase, ERK: extracellular-signal-regulated kinase.
To this end, the physiological/pathological relevance of CRABP1 is most evident in cancers. For instance, the CRABP1 gene has been reported as a tumor suppressor or an oncogene in animals and humans [15][49][50][51][52][53][54][55][56][57][58][59][60][61]. In comparing CKO and wild-type ESCs, as well as in gain- and loss-of-functional studies of cancer cell models, it was found that CRABP1 was involved in modulating cell cycle control [22]. By competing with Ras for forming complexes with RAF/MEK, atRA-CRABP1 dampened mitogen-activated ERK activity and suppressed cell cycle progression by expanding the G1 phase [22][29]. This supports the notion that CRABP1 can be a tumor suppressor. Additional evidence supporting a functional role for CRABP1 in stem cell proliferation was obtained from studying CKO mice that were found to have expanded NSC pools (as a result of enhanced NSC proliferation in CRABP1-deleted hippocampus), which was consistent with the CKO mouse behavior indicating improved memory function [25]. Importantly, the hippocampus is among the tissues where CRABP1 is most highly expressed, especially in the NSC-rich region of the dentate gyrus. Thus, CRABP1 can participate in the homeostatic control of the NSC pool in the brain. Readers are referred to an in-depth review of this CRABP1-regulated signaling pathway by Nagpal and Wei [62].
2.2. Crabp1-MAPK Signalsome in Metabolism and Immunity
Lin et al. first observed that CKO mice exhibited increased high-fat diet (HFD)-induced obesity and insulin resistance (IR), suggesting a protective role for CRABP1 against the development of metabolic disorders. A molecular study of CKO mice elucidated an underlying mechanism for this metabolic phenotype that, in normal adipocytes, CRABP1 could negatively regulate ERK activity to inhibit adipogenesis and adipose hypertrophy [28]. Therefore, CKO mice are more prone to HFD-induced obesity and IR. To this end, it has been reported that pharmacological doses of RA could inhibit adipogenesis and protect against obesity, and this was attributed, primarily, to RAR-mediated activities [63][64][65][66][67]. These recent studies of CKO models revealed CRABP1 as an additional player in mediating physiological activities of atRA regarding metabolic homeostasis and the maintenance of healthy adipose tissue [28].
In examining the systemic inflammatory status/potential of CKO mice, it was found that HFD-fed CKO mice all had increased systemic inflammation, indicated by invading immune cells in adipose tissue [28], increased inflammatory driver Receptor Interacting Protein 140 (RIP140) (gene name Nrip1) [68] in the blood [31], elevation in inflammatory cytokines, and significantly enhanced macrophage M1 polarization (unpublished). Previous studies also indicated that CKO mice had overall increased inflammation in the heart, indicated by increased cardiac fibrosis [26], and an altered anxiety and stress response in their HPA axis [32]. To this end, CRABP1 was found to be involved in exosome secretion from CRABP1-expressing neurons. Specifically, the RIP140-containing exosome population was significantly expanded in the blood and cerebral spinal fluid (CSF) of CKO mice, due to, in part, increased exosome secretion from CKO neurons [31]. Importantly, these neuron-derived RIP140-containing exosomes could be engulfed by macrophages to increase their inflammatory M1 polarization, thereby increasing systemic inflammation. This study, by monitoring the intercellular transfer of the inflammatory driver, RIP140, demonstrates exosome secretion as a potent means to transfer neuronal inflammation into systemic inflammation; mechanistically, this study identifies CRABP1 as an important regulator of exosome secretion from specific CRABP1-expressing neurons, which also involves the MAPK-ERK signaling in these neurons [31].
2.3. CRABP1-CaMKII Signalsome in Cardiomyocytes and Motor Neurons (MNs)
A different CRABP1-signaling complex was identified from studying deteriorated heart function of CKO mice [26][27], and their premature weakening in motor function [33]. The expression study confirmed CRABP1 expression in cardiomyocytes [26] (relevant to the CKO heart phenotype) and motor neurons (relevant to the CKO motor function phenotype) [33]. This signaling complex is comprised of CRABP1 and calcium-calmodulin-dependent kinase 2 (CaMKII), an enzyme critical to calcium signaling/handling and highly enriched in both cardiomyocytes [69] and neurons [70][71]. It is known that CaMKII regulates contraction in cardiomyocytes [69] and long-term potentiation in neurons [70][71], respectively. Both types of cells are highly dependent upon calcium homeostasis for their functions where CaMKII is a key mediator of calcium signaling [72]. All the CaMKII isoforms have a conserved architecture comprised of the kinase, regulatory, and association/oligomerization domains, and share the same activation mechanism through the binding of calmodulin to the calmodulin-binding domain (CaMBD) within its regulatory domain. CaMKII activation occurs when intracellular (Ca2+) increases and binds calmodulin. Ca2+-calmodulin then binds and activates CaMKII, which is often marked by phosphorylation at threonine 286/7 (T286/7), depending on the CaMKII isoform [73][74]. In vitro data showed that CRABP1competes with calmodulin by directly interacting with CaMKII at the CaMBD [26][27]. Therefore, CRABP1 could dampen Ca2+/Calmodulin activated CaMKII activity. Since over-activation of CaMKII is a major trigger of the death/damage of cardiomyocytes [75] and neurons [76], by dampening CaMKII over-activation, CRABP1 can play a protective role in maintaining the health of both the heart and neurons. These are elaborated on in the following section. The proposed mechanistic model for CRABP1-CaMKII signalsome is shown in Figure 3.
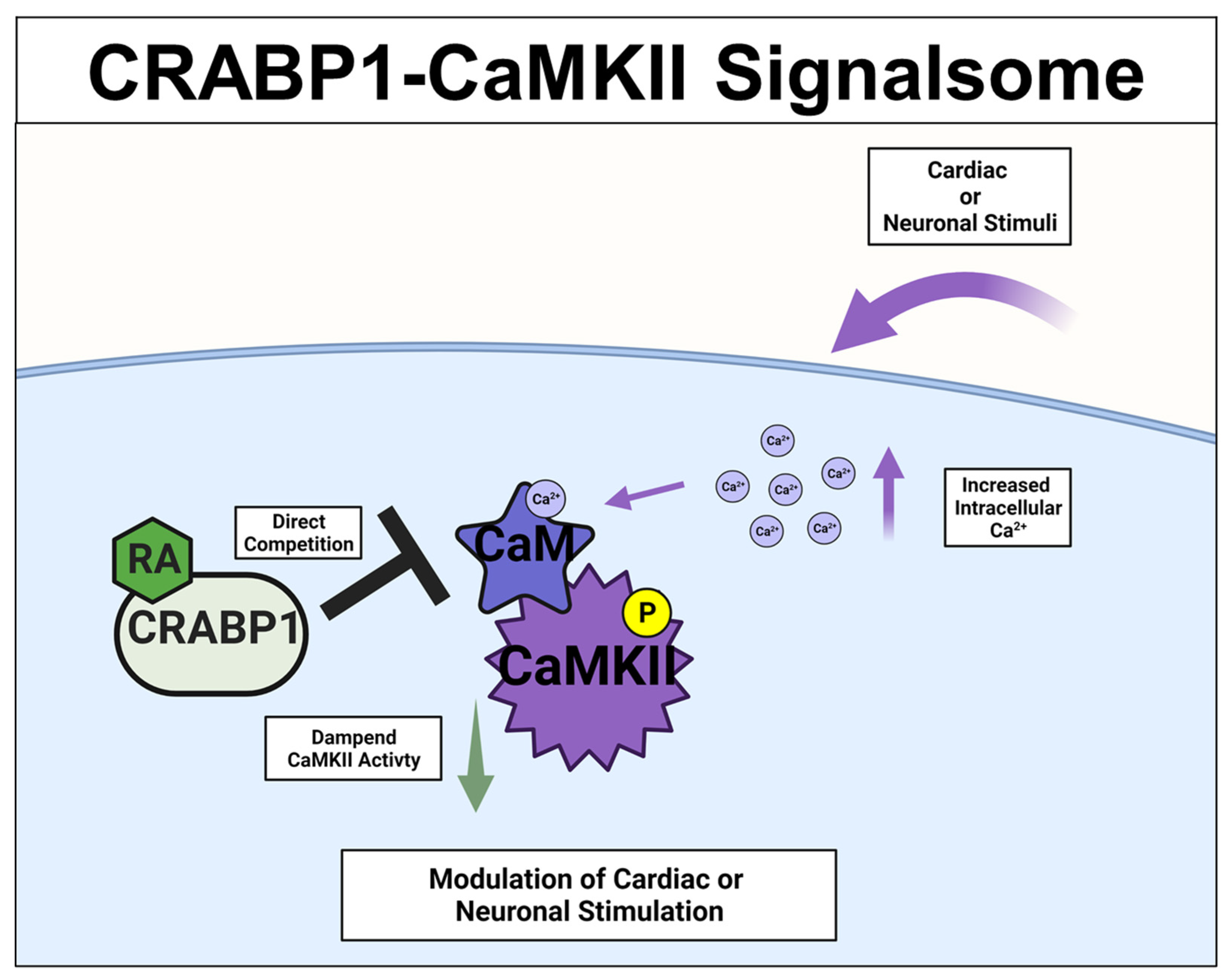
Figure 3. CRABP1-CaMKII signalsome. Upon cardiac or neuronal stimulation and subsequent intracellular Ca2+ increase to activate CaMKII, CRABP1 directly competes with calmodulin (CaM) to dampen CaMKII enzyme activity to ultimately modulate cardiac and/or neuronal stimulation. CRABP1: Cellular Retinoic Acid Binding Protein 1, RA: retinoic acid, CaMKII: calcium-calmodulin-associated dependent kinase 2.
References
- Tanumihardjo, S.A.; Russell, R.M.; Stephensen, C.B.; Gannon, B.M.; Craft, N.E.; Haskell, M.J.; Lietz, G.; Schulze, K.; Raiten, D.J. Biomarkers of nutrition for development (BOND)-vitamin A review. J. Nutr. 2016, 146, 1816S–1848S.
- Cunningham, T.J.; Duester, G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015, 16, 110–123.
- Duester, G. Retinoic acid synthesis and signaling during early organogenesis. Cell 2008, 134, 921–931.
- Wolf, G. Retinoic acid as cause of cell proliferation or cell growth inhibition depending on activation of one of two different nuclear receptors. Nutr. Rev. 2008, 66, 55–59.
- Gudas, L.J.; Wagner, J.A. Retinoids regulate stem cell differentiation. J. Cell. Physiol. 2011, 226, 322–330.
- Noy, N. Between death and survival: Retinoic acid in regulation of apoptosis. Annu. Rev. Nutr. 2010, 30, 201–217.
- Niederreither, K.; Dollé, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553.
- Rothman, K.J.; Moore, L.L.; Singer, M.R.; Nguyen, U.-S.D.T.; Mannino, S.; Milunsky, A. Teratogenicity of High Vitamin A Intake. N. Engl. J. Med. 1995, 333, 1369–1373.
- Shenefelt, R.E. Morphogenesis of malformations in hamsters caused by retinoic acid: Relation to dose and stage at treatment. Teratology 5:103-18. 1972. Birth Defects Res. A Clin. Mol. Teratol. 2010, 88, 847–862.
- Wilson, J.G.; Roth, C.B.; Warkany, J. An analysis of the syndrome of malformations induced by maternal vitamin a deficiency. Effects of restoration of vitamin a at various times during gestation. Am. J. Anat. 1953, 92, 189–217.
- Duong, V.; Rochette-Egly, C. The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 1023–1031.
- Topletz, A.R.; Thatcher, J.E.; Zelter, A.; Lutz, J.D.; Tay, S.; Nelson, W.L.; Isoherranen, N. Comparison of the function and expression of CYP26A1 and CYP26B1, the two retinoic acid hydroxylases. Biochem. Pharmacol. 2012, 83, 149–163.
- Topletz, A.R.; Tripathy, S.; Foti, R.S.; Shimshoni, J.A.; Nelson, W.L.; Isoherranen, N. Induction of CYP26A1 by metabolites of retinoic acid: Evidence that CYP26A1 is an important enzyme in the elimination of active retinoids. Mol. Pharmacol. 2015, 87, 430–441.
- Napoli, J.L. Functions of intracellular retinoid binding-proteins. Subcell. Biochem. 2016, 81, 21–76.
- Napoli, J.L. Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: Effects on retinoid metabolism, function and related diseases. Pharmacol Ther 2017, 173, 19–33.
- Stevison, F.; Jing, J.; Tripathy, S.; Isoherranen, N. Role of Retinoic Acid-Metabolizing Cytochrome P450s, CYP26, in Inflammation and Cancer. Adv. Pharmacol. 2015, 74, 373–412.
- Thatcher, J.E.; Isoherranen, N. The role of CYP26 enzymes in retinoic acid clearance. Expert Opin. Drug Metab. Toxicol. 2009, 5, 875–886.
- Dong, D.; Ruuska, S.E.; Levinthal, D.J.; Noy, N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 1999, 274, 23695–23698.
- Majumdar, A.; Petrescu, A.D.; Xiong, Y.; Noys, N. Nuclear translocation of cellular retinoic acid-binding protein II is regulated by retinoic acid-controlled SUMOylation. J. Biol. Chem. 2011, 286, 42749–42757.
- Gupta, P.; Ho, P.-C.; Huq, M.M.; Ha, S.G.; Park, S.W.; Khan, A.A.; Tsai, N.-P.; Wei, L.-N. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc. Natl. Acad. Sci. USA 2008, 105, 11424–11429.
- Chuang, Y.-S.S.; Huang, W.-H.H.; Park, S.W.; Persaud, S.D.; Hung, C.-H.H.; Ho, P.-C.C.; Wei, L.-N.N. Promyelocytic leukemia protein in retinoic acid-induced chromatin remodeling of Oct4 gene promoter. Stem Cells 2011, 29, 660–669.
- Persaud, S.D.; Lin, Y.-W.; Wu, C.-Y.; Kagechika, H.; Wei, L.-N. Cellular retinoic acid binding protein I mediates rapid non-canonical activation of ERK1/2 by all-trans retinoic acid. Cell. Signal. 2013, 25, 19–25.
- Wu, C.Y.; Persaud, S.D.; Wei, L.N. Retinoic Acid Induces Ubiquitination-Resistant RIP140/LSD1 Complex to Fine-Tune Pax6 Gene in Neuronal Differentiation. Stem Cells 2016, 34, 114–123.
- Persaud, S.D.; Park, S.W.; Ishigami-Yuasa, M.; Koyano-Nakagawa, N.; Kagechika, H.; Wei, L.N. All trans-retinoic acid analogs promote cancer cell apoptosis through non-genomic Crabp1 mediating ERK1/2 phosphorylation. Sci. Rep. 2016, 6, 22396.
- Lin, Y.L.; Persaud, S.D.; Nhieu, J.; Wei, L.N. Cellular Retinoic Acid-Binding Protein 1 Modulates Stem Cell Proliferation to Affect Learning and Memory in Male Mice. Endocrinology 2017, 158, 3004–3014.
- Park, S.W.; Persaud, S.D.; Ogokeh, S.; Meyers, T.A.; Townsend, D.W.; Wei, L.N. CRABP1 protects the heart from isoproterenol-induced acute and chronic remodeling. J. Endocrinol. 2018, 236, 151–165.
- Park, S.W.; Nhieu, J.; Lin, Y.W.; Wei, L.N. All-trans retinoic acid attenuates isoproterenol-induced cardiac dysfunction through Crabp1 to dampen CaMKII activation. Eur. J. Pharmacol. 2019, 858, 172485.
- Lin, Y.W.; Park, S.W.; Lin, Y.L.; Burton, F.H.; Wei, L.N. Cellular retinoic acid binding protein 1 protects mice from high-fat diet-induced obesity by decreasing adipocyte hypertrophy. Int. J. Obes. 2020, 44, 466–474.
- Wook Park, S.; Nhieu, J.; Persaud, S.D.; Miller, M.C.; Xia, Y.; Lin, Y.W.; Lin, Y.L.; Kagechika, H.; Mayo, K.H.; Wei, L.N. A new regulatory mechanism for Raf kinase activation, retinoic acid-bound Crabp1. Sci. Rep. 2019, 9, 10929.
- Lin, Y.W.Y.L.W.; Nhieu, J.; Zhang, X.; Wei, L.N. Sonic hedgehog-gli1 signaling and cellular retinoic acid binding protein 1 gene regulation in motor neuron differentiation and diseases. Int. J. Mol. Sci. 2020, 21, 4125.
- Lin, Y.W.; Nhieu, J.; Wei, C.W.; Lin, Y.L.; Kagechika, H.; Wei, L.N. Regulation of exosome secretion by cellular retinoic acid binding protein 1 contributes to systemic anti-inflammation. Cell Commun. Signal. 2021, 19, 69.
- Lin, Y.L.; Wei, C.W.; Lerdall, T.A.; Nhieu, J.; Wei, L.N. Crabp1 modulates hpa axis homeostasis and anxiety-like behaviors by altering fkbp5 expression. Int. J. Mol. Sci. 2021, 22, 12240.
- Lin, Y.-L.Y.-W.; Nhieu, J.; Liu, P.-Y.; Le, G.; Lee, D.J.; Wei, C.-W.; Lin, Y.-L.Y.-W.; Oh, S.-H.; Lowe, D.; Wei, L.-N. CRABP1-CaMKII-Agrn regulates the maintenance of neuromuscular junction in spinal motor neuron. Cell Death Differ. 2022, 1–13.
- Ong, D.E.; Chytil, F. Cellular retinoic acid-binding protein from rat testis. Purification and characterization. J. Biol. Chem. 1978, 253, 4551–4554.
- Fiorella, P.D.; Giguère, V.; Napoli, J.L. Expression of cellular retinoic acid-binding protein (type II) in Escherichia coli: Characterization and comparison to cellular retinoic acid-binding protein (type I). J. Biol. Chem. 1993, 268, 21545–21552.
- Norris, A.W.; Cheng, L.; Giguère, V.; Rosenberger, M.; Li, E. Measurement of subnanomolar retinoic acid binding affinities for cellular retinoic acid binding proteins by fluorometric titration. Biochim. Biophys. Acta 1994, 1209, 10–18.
- Wang, L.; Li, Y.; Yan, H. Structure-function relationships of cellular retinoic acid-binding proteins: Quantitative analysis of the ligand binding properties of the wild-type proteins and site-directed mutants. J. Biol. Chem. 1997, 272, 1541–1547.
- Wei, L.N.; Chang, L.; Lee, C.H. Studies of over-expressing cellular retinoic acid binding protein-I in cultured cells and transgenic mice. Transgenics 1997, 2, 201–209.
- Wei, L.N.; Chang, L.; Hu, X. Studies of the type I cellular retinoic acid-binding protein mutants and their biological activities. Mol. Cell. Biochem. 1999, 200, 69–76.
- Nelson, C.H.; Peng, C.C.; Lutz, J.D.; Yeung, C.K.; Zelter, A.; Isoherranen, N. Direct protein–protein interactions and substrate channeling between cellular retinoic acid binding proteins and CYP26B1. FEBS Lett. 2016, 590, 2527–2535.
- Eller, M.S.; Oleksiak, M.F.; McQuaid, T.J.; McAfee, S.G.; Gilchrest, B.A. The molecular cloning and expression of two CRABP cDNAs from human skin. Exp. Cell Res. 1992, 198, 328–336.
- Tang, Z.; Li, Y.; Wan, P.; Li, X.; Zhao, S.; Liu, B.; Fan, B.; Zhu, M.; Yu, M.; Li, K. LongSAGE analysis of skeletal muscle at three prenatal stages in Tongcheng and Landrace pigs. Genome Biol. 2007, 8, R115.
- Chapman, J.M.; Curley, R.W. Affinity purification of retinoic acid-binding proteins using immobilized 4-(2-Hydroxyethoxy)retinoic acid. Protein Expr. Purif. 1990, 1, 63–69.
- Stoner, C.M.; Gudas, L.J. Mouse Cellular Retinoic Acid Binding Protein: Cloning, Complementary DNA Sequence, and Messenger RNA Expression during the Retinoic Acid-induced Differentiation of F9 Wild Type and RA-3-10 Mutant Teratocarcinoma Cells. Cancer Res. 1989, 49, 1497–1504.
- Nilsson, M.H.L.; Spurr, N.K.; Saksena, P.; Busch, C.; Nordlinder, H.; Peterson, P.A.; Rask, L.; Sundelin, J. Isolation and characterization of a cDNA clone corresponding to bovine cellular retinoic-acid-binding protein and chromosomal localization of the corresponding human gene. Eur. J. Biochem. 1988, 173, 45–51.
- Wei, L.-N. Non-canonical activity of retinoic acid in epigenetic control of embryonic stem cell. Transcription 2013, 4, 158–161.
- Wei, L.N. Cellular retinoic acid binding proteins: Genomic and non-genomic functions and their regulation. Subcell. Biochem. 2016, 81, 163–178.
- Zhang, W.; Liu, H.T.; LIU, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18.
- Guidez, F.; Parks, S.; Wong, H.; Jovanovic, J.V.; Mays, A.; Gilkes, A.F.; Mills, K.I.; Guillemin, M.-C.C.; Hobbs, R.M.; Pandolfi, P.P.; et al. RARalpha-PLZF overcomes PLZF-mediated repression of CRABPI, contributing to retinoid resistance in t(11;17) acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 18694–18699.
- Pfoertner, S.; Goelden, U.; Hansen, W.; Toepfer, T.; Geffers, R.; Ukena, S.N.; von Knobloch, R.; Hofmann, R.; Buer, J.; Schrader, A.J. Cellular retinoic acid binding protein I: Expression and functional influence in renal cell carcinoma. Tumour Biol. 2005, 26, 313–323.
- Tanaka, K.; Imoto, I.; Inoue, J.; Kozaki, K.; Tsuda, H.; Shimada, Y.; Aiko, S.; Yoshizumi, Y.; Iwai, T.; Kawano, T.; et al. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene 2007, 26, 6456–6468.
- Miyake, T.; Ueda, Y.; Matsuzaki, S.; Miyatake, T.; Yoshino, K.; Fujita, M.; Nomura, T.; Enomoto, T.; Kimura, T. CRABP1-reduced expression is associated with poorer prognosis in serous and clear cell ovarian adenocarcinoma. J. Cancer Res. Clin. Oncol. 2011, 137, 715–722.
- Wu, Q.; Lothe, R.A.; Ahlquist, T.; Silins, I.; Tropé, C.G.; Micci, F.; Nesland, J.M.; Suo, Z.; Lind, G.E. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol. Cancer 2007, 6, 45.
- Hawthorn, L.; Stein, L.; Varma, R.; Wiseman, S.; Loree, T.; Tan, D.F. TIMP1 and SERPIN-A overexpression and TFF3 and CRABP1 underexpression as biomarkers for papillary thyroid carcinoma. Head Neck 2004, 26, 1069–1083.
- Celestino, R.; Nome, T.; Pestana, A.; Hoff, A.M.; Gonçalves, A.P.; Pereira, L.; Cavadas, B.; Eloy, C.; Bjøro, T.; Sobrinho-Simões, M.; et al. CRABP1, C1QL1 and LCN2 are biomarkers of differentiated thyroid carcinoma, and predict extrathyroidal extension. BMC Cancer 2018, 18, 68.
- Huang, Y.; de la Chapelle, A.; Pellegata, N.S. Hypermethylation, but not LOH, is associated with the low expression of MT1G and CRABP1 in papillary thyroid carcinoma. Int. J. Cancer 2003, 104, 735–744.
- Lind, G.E.; Kleivi, K.; Meling, G.I.; Teixeira, M.R.; Thiis-Evensen, E.; Rognum, T.O.; Lothe, R.A. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Cell. Oncol. 2006, 28, 259–272.
- Won, J.Y.; Nam, E.C.; Yoo, S.J.; Kwon, H.J.; Um, S.J.; Han, H.S.; Kim, S.H.; Byun, Y.; Kim, S.Y. The effect of cellular retinoic acid binding protein-I expression on the CYP26-mediated catabolism of all-trans retinoic acid and cell proliferation in head and neck squamous cell carcinoma. Metabolism 2004, 53, 1007–1012.
- Kainov, Y.; Favorskaya, I.; Delektorskaya, V.; Chemeris, G.; Komelkov, A.; Zhuravskaya, A.; Trukhanova, L.; Zueva, E.; Tavitian, B.; Dyakova, N.; et al. CRABP1 provides high malignancy of transformed mesenchymal cells and contributes to the pathogenesis of mesenchymal and neuroendocrine tumors. Cell Cycle 2014, 13, 1530–1539.
- Choi, N.; Park, J.; Lee, J.S.; Yoe, J.; Park, G.Y.; Kim, E.; Jeon, H.; Cho, Y.M.; Roh, T.Y.; Lee, Y. miR-93/miR-106b/miR-375-CIC-CRABP1: A novel regulatory axis in prostate cancer progression. Oncotarget 2015, 6, 23533–23547.
- Liu, R.Z.; Garcia, E.; Glubrecht, D.D.; Poon, H.Y.; Mackey, J.R.; Godbout, R. CRABP1 is associated with a poor prognosis in breast cancer: Adding to the complexity of breast cancer cell response to retinoic acid. Mol. Cancer 2015, 14, 129.
- Nagpal, I.; Wei, L.N. All-trans retinoic acid as a versatile cytosolic signal modulator mediated by CRABP1. Int. J. Mol. Sci. 2019, 20, 3610.
- Berry, D.C.; DeSantis, D.; Soltanian, H.; Croniger, C.M.; Noy, N. Retinoic acid upregulates preadipocyte genes to block adipogenesis and suppress diet-induced obesity. Diabetes 2012, 61, 1112–1121.
- Berry, D.C.; Noy, N. All-trans-Retinoic Acid Represses Obesity and Insulin Resistance by Activating both Peroxisome Proliferation-Activated Receptor β/δ and Retinoic Acid Receptor. Mol. Cell. Biol. 2009, 29, 3286–3296.
- Jeyakumar, S.M.; Vajreswari, A.; Sesikeran, B.; Giridharan, N.V. Vitamin A supplementation induces adipose tissue loss through apoptosis in lean but not in obese rats of the WNIN/Ob strain. J. Mol. Endocrinol. 2005, 35, 391–398.
- Kamei, Y.; Kawada, T.; Mizukami, J.; Sugimoto, E. The prevention of adipose differentiation of 3T3-L1 cells caused by retinoic acid is elicited through retinoic acid receptor alpha. Life Sci. 1994, 55, PL307–PL312.
- Murray, T.; Russell, T.R. Inhibition of adipose conversion in 3T3-L2 cells by retinoic acid. J. Supramol. Cell. Biochem. 1980, 14, 255–266.
- Lee, B.; Ho, P.-C.; Wei, L.-N. Nuclear Receptor-Interacting Protein 1 (NRIP1). In Encyclopedia of Signaling Molecules; Springer: Cham, Switzerland, 2018; pp. 3606–3616.
- Erickson, J.R. Mechanisms of CaMKII activation in the heart. Front. Pharmacol. 2014, 5 APR, 59.
- Lisman, J.; Schulman, H.; Cline, H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 2002, 3, 175–190.
- Lisman, J.; Yasuda, R.; Raghavachari, S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012, 13, 169–182.
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21.
- Bhattacharyya, M.; Karandur, D.; Kuriyan, J. Structural insights into the regulation of Ca2+/calmodulin-dependent protein kinase II (Camkii). Cold Spring Harb. Perspect. Biol. 2020, 12, 1–20.
- Stratton, M.M.; Chao, L.H.; Schulman, H.; Kuriyan, J. Structural studies on the regulation of Ca2+/calmodulin dependent protein kinase II. Curr. Opin. Struct. Biol. 2013, 23, 292–301.
- Zhang, P. CaMKII: The molecular villain that aggravates cardiovascular disease. Exp. Ther. Med. 2017, 13, 815–820.
- Ashpole, N.M.; Hudmon, A. Excitotoxic neuroprotection and vulnerability with CaMKII inhibition. Mol. Cell. Neurosci. 2011, 46, 720–730.
More
Information
Subjects:
Medicine, Research & Experimental
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Entry Collection:
Biopharmaceuticals Technology
Revisions:
3 times
(View History)
Update Date:
21 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




