Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Adam E. Frampton | -- | 1695 | 2022-04-20 11:35:13 | | | |
| 2 | Beatrix Zheng | + 5 word(s) | 1700 | 2022-04-21 04:28:30 | | | | |
| 3 | Daniel Liu | Meta information modification | 1700 | 2022-04-21 09:32:08 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Frampton, A.; Liu, D.; , .; Asim, M. Transfer RNAs Present in Extracellular Vesicles. Encyclopedia. Available online: https://encyclopedia.pub/entry/21999 (accessed on 08 February 2026).
Frampton A, Liu D, , Asim M. Transfer RNAs Present in Extracellular Vesicles. Encyclopedia. Available at: https://encyclopedia.pub/entry/21999. Accessed February 08, 2026.
Frampton, Adam, Daniel Liu, , Mohammad Asim. "Transfer RNAs Present in Extracellular Vesicles" Encyclopedia, https://encyclopedia.pub/entry/21999 (accessed February 08, 2026).
Frampton, A., Liu, D., , ., & Asim, M. (2022, April 20). Transfer RNAs Present in Extracellular Vesicles. In Encyclopedia. https://encyclopedia.pub/entry/21999
Frampton, Adam, et al. "Transfer RNAs Present in Extracellular Vesicles." Encyclopedia. Web. 20 April, 2022.
Copy Citation
Extracellular vesicles (EVs) are small cargo-containing structures with a lipid bilayer but do not have the cellular machinery required to replicate. They have been shown to play a role in cell-to-cell communication, as they can be found to transport biological material including proteins, lipids, ribonucleic acid (RNA), and deoxyribonucleic acid (DNA) between cells, leading to cellular changes within multi-cellular organisms. It is a process that has been conserved through evolution, found both in prokaryotes and eukaryotes.
extracellular vesicle
exosome
transfer RNA
tRNA fragment
tRNA half
cancer
1. Introduction
Extracellular vesicles (EVs) are an umbrella term for a heterogenous array of secreted membrane vesicles and can be further distinguished by cell source, biogenesis pathways, and size ranges, thus contributing to diverse nomenclature [1]. Two broad categories that are often used distinguish microvesicles (MVs) and exosomes on the basis of cellular biogenesis [1]. Microvesicles are generated by the outward budding of the phospholipid bilayer generating vesicles made directly from the plasma membrane. These vesicles tend to be larger with MVs having a size range of up to 1000 nm or larger in some cases [1][2]. Exosomes, on the other hand, involve intracellular mechanisms forming multivesicular endosomes (MVEs) which contain intraluminal vesicles (ILVs). ILVs are formed from the budding of MVEs which then fuse with the plasma membrane and get released extracellularly as exosomes. Exosomes are usually smaller in size (<150 nm) closely reflecting that of ILVs [1][2] and therefore also express specific markers that relate to their endosomal origin [1].
EVs have been found to be present in virtually all types of biofluids, including cerebrospinal fluid (CSF) [3], urine [4], blood [5], bile [6], and breast milk [7] as well as being released by other organisms and even plants [8]. Numerous studies focus on their carrier material which contains lipids, protein, and RNA in an endeavour to link them to the pathophysiology of a wide range of diseases [9]. With the current advances in RNA sequencing technologies and bioinformatic strategies, this has drawn attention to the wide range of small RNA (sRNA) content in EVs.
Whilst much of the literature has focused on studying micro RNAs (miRNAs) [10], short 16–22 nucleotide (nt) single-stranded molecules with a role in canonical transcription and translational pathways, less is known about the role of transfer RNAs (tRNA) despite being the most abundant RNA in the human genome [11], and the first non-coding RNA to have been discovered [12]. The canonical cytoplasmic tRNA molecule is 76 nt long and well-conserved, with a cloverleaf secondary structure and a tertiary L-shaped structure that separates the anticodon triplet for recognition of template mRNA from the amino acid attachment site. Whilst tRNAs are present in all species for protein translation, tRNA genetic expression is species-specific and with over 270 different tRNA sequences present among approximately 450 tRNA human genes, highly complex in eukaryotes [13]. Furthermore, the genetic expression of tRNAs is subject to modifications such as base-specific methylation, altering function, and fragmentation which can generate whole new subspecies of RNA molecules. This has led to, over the past decade [14][15], a wide expansion in tRNA biology with regulatory functions affecting gene expression, protein synthesis, and the stress response as significant downstream functions [11].
The fragmentation products of tRNAs are a potential source of new functionality away from conventional protein synthesis. tRNA-halves and tRNA-derived fragments (tRFs) are two such fragmentation products of tRNAs and have distinct biogenesis pathways (Figure 1). tRNA-halves are thought to be formed by nucleases such as angiogenin as well as Dicer and RNase Z which are upregulated under varying cellular conditions including stress [11][14][16]. The simplest fragmentation is a cleavage of the anti-codon loop forming either 5′ or 3′ tRNA-halves with lengths corresponding to half the full-length of mature tRNAs [14]. tRFs on the other hand, includes 5′ and 3′-tRFs with shorter nucleotide sequences and are named based on where the cleavage occurred, and are thought to be produced from the cleavage occurring in or around the D-loop or T-loop structure of mature tRNAs, respectively [16]. However, tRFs can also arise from 3′ nuclease cleavage of 5′ tRNA-halves and as such, the enzymes responsible for the production of smaller tRFs remain not well understood [16]. There are also tRFs that do not correspond to the 5′ or 3′ end and are called internal tRFs. Downstream effects of tRFs are thought to be related to RNA machinery, associating with argonaute proteins to exhibit miRNA-like regulatory and silencing activity on mRNA to facilitate post-transcriptional repression [16]. This represents a new spectrum of molecular pathways that converge to modify gene expression and thus influence cells in health and disease.
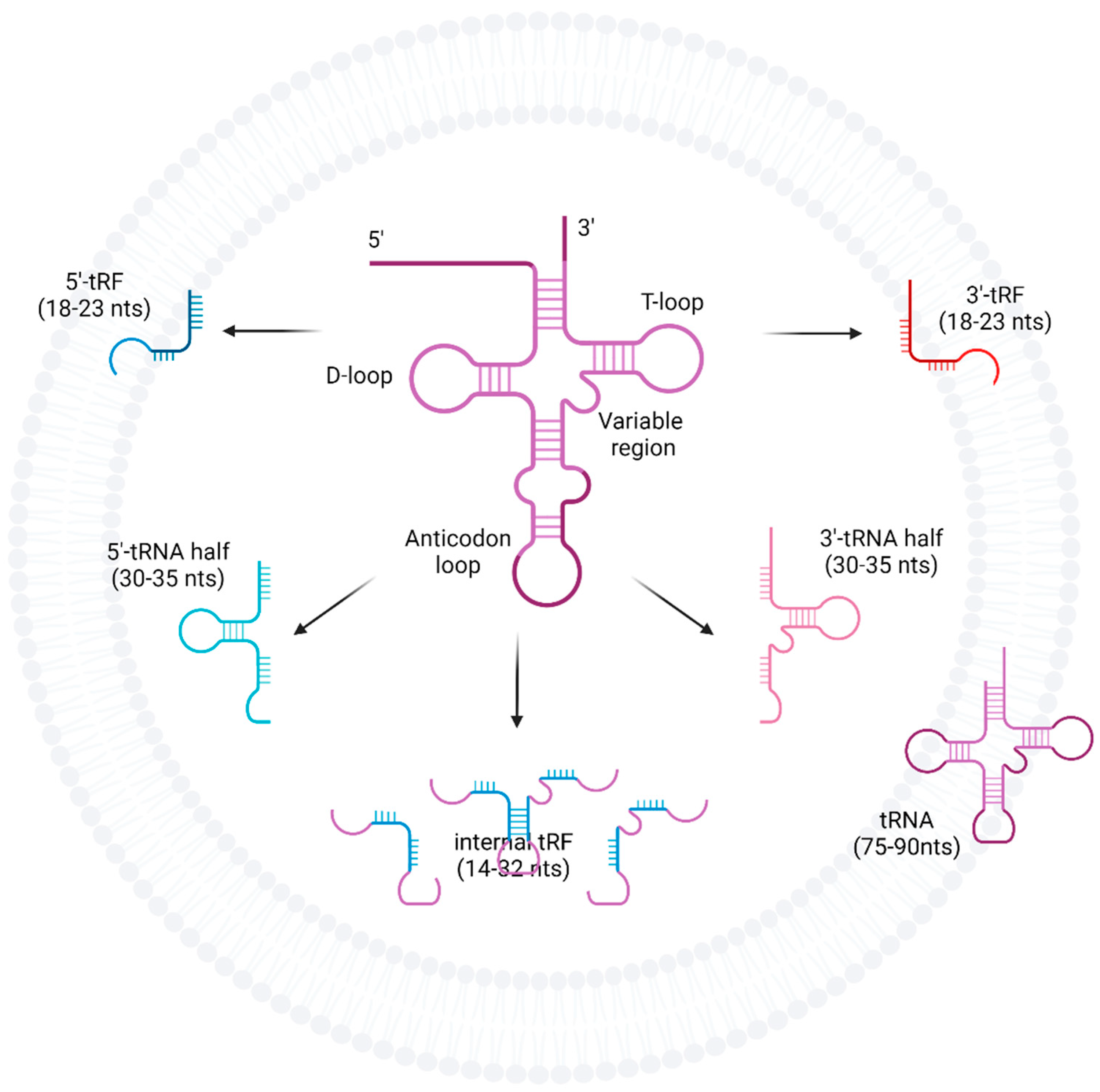
Figure 1. Fragmentation of tRNAs into tRFs, tRNA-halves, and internal tRFs. tRNAs are cleaved at the anti-codon loop by enzymes such as angiogenin, Dicer, or RNase Z into either 5′ or 3′-tRNA-halves. tRFs can arise from mature tRNA, pre-tRNA as well as tRNA-halves, and are formed when cleavage occurs at either the D-loop or T-loop. However, the enzymes responsible for tRF production are less well understood.
2. Current Insights
The researchers have focussed on the tRNAs, tRNA-halves, and tRFs associated with human EVs, in health and pathological disease states. They have described the various biofluids used for EV capture and characterised the different EV isolation methods used in protocols for the subsequent identification of EV tRNAs contained within. Importantly, the researchers have shown that multiple pathologies and especially malignancies can be distinguished by differential EV tRNA expression and may thus be of clinical significance for both diagnosis and prognosis of the disease.
The abundance of tRNAs contained within EVs is highly variable and may depend on tissue and disease type, ranging from as low as 0.04% in the blood plasma EVs of gastric cancer patients [17] to as high as >95% from EVs obtained from healthy placental syncytiotrophoblasts [18]. However, tRNAs appear to be a consistent finding in EVs, occurring in sequencing samples with as high a prevalence (and sometimes higher) as miRNAs. Additionally, EV isolation methods will need to be tailored to the requirements of the experiment, with highly selective methods such as density gradient preferable when contamination with RBP is to be avoided. The researchers found that 46.5% of the protocols used precipitation as their EV isolation method, and this has been recognised as having relatively low purity [19][20]. Moreover, the technological limitations of sequencing methods may further add to the complexity of interpreting the EV tRNAs found. The research highlighted a few limitations of current sequencing technologies. In many studies, the library preparation is limited to specific size ranges [21] and low abundance species can lead to selective biases in enrichment. [22] Moreover, conventional sequencing methods may not effectively capture RNAs that are post-transcriptionally modified [23]. Finally, Tosar et al. [24] highlighted how piRNA transcripts differ from some tRNA-halves by only one nucleotide. Individual piRNA and as well as other small cytoplasmic RNA species are highly homologous to major tRNA fragments within commonly used databases; thus, mapping of reads may lead to an under- or over-representation of tRNA fragments. The limitations encountered with sequencing technologies could be addressed through novel pipelines such as that reported by Amorim et al. [25]. Ongoing collaborative work such as the Extracellular RNA Communication Consortium has made efforts to improve this and work towards strategies such as deconvolution to determine tRNA sources [26].
It is increasingly recognised that tRNA-derived fragments have a role in mammalian cells, with 5′-tRNA-halves having been associated with actions on the ribosome leading to stress granule formation [27] and translational inhibition. Smaller tRFs share a size similarity to miRNAs and there is significant evidence in the wider (non-EV) literature that they can have actions in conjunction with Argonaute [28], the Piwi subfamily, and other RNA-binding proteins [28][29].
tRNAs and their fragmentation products have been found as an EV biomarker in circulating fluid and tissue for a number of malignancies including leukaemia, prostate, ovarian, pancreatic, and colorectal cancers [30][31]. In the research, breast cancer showed particular promise with several differentially expressed tRNAs (tRF-Arg-CCT-017, tRF-Gly-CCC-001, and tiRNA-Phe-GAA-003) associated with key regulatory pathways such as Wnt signalling, classically involved in cancer initiation and progression. [32] In fact, several other malignancies such as colorectal cancers [33], gastric cancers [34], and glioblastomas [35] also secrete cancer-stem-cell-associated EVs, which act on the β-catenin/Wnt-signalling pathways to increase stemness, in turn increasing their tumorigenic potential.
In addition, the majority of the studies in this research showed that the fragmentation products of tRNAs, namely 5′-tRFs and 5′ tRNA-halves, were the most abundant sub-species of tRNAs identified in EVs. A possible reason for the observation of 5′ products was shown to be potentially due to reverse transcription failure due to post-transcriptional modifications in otherwise full-length tRNAs by Shurtleff et al. who used an alternative thermostable sequencing system. What is also clear is that simple analysis of tRNA molecules by broad amino acid (isoacceptor types) may not be sophisticated enough to determine potentially clinically relevant changes in EV tRNA profiles. Further validation of sequencing by qRT-PCR is necessary to prove true differential expression and this means additionally incorporating the standardised use of tRNA molecular terminology.
The researchers have also highlighted that several biological mechanisms have been clearly associated with changes in tRNA-halves levels. For instance, osteoblastogenesis revealed that BMSCs alter their cargo content in EVs from full-length tRNAs to tRNA-halves as their differentiation progresses [36]. The researchers have noted the evidence that there are time-dependent changes in the EV tRNA profile of cancer patients and may perhaps reflect clinical changes such as disease burden [37]. In addition, tRFs have even been shown to possess potential diagnostic potential in a multitude of diseases.
tRNA-sorting mechanisms in EVs have also been the focus of various studies, with RNA-binding proteins [23], surface markers [38], and tRNA enzymatic stability as some of the mechanisms identified [39]. Potential therapeutic uses of EV tRNA include the loading of synthetic oligonucleotides into EVs via parent cells to alter downstream gene expression in donor cells [39]. This particular paper by Gambaro et al. [39] demonstrated key mechanisms for tRNA sorting, such as the concentration-dependent transfection and selection of stable synthetic oligonucleotides against enzyme degradation, through well-controlled transfection experiments.
Due to the heterogeneity of the studies included in this research, a meta-analysis could not be conducted; thus, a narrative synthesis of the literature was undertaken. Many studies may include tRNA data but not explicitly mention this and will have been excluded. The researchers also restricted their search to articles involving human cell models or patient-derived biofluids which may exclude important studies using animal EV models that could have shed some light on tRNA markers across other species.
References
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Moller, A.; Lobb, R.J. The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer 2020, 20, 697–709.
- Welton, J.L.; Loveless, S.; Stone, T.; von Ruhland, C.; Robertson, N.P.; Clayton, A. Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis. J. Extracell. Vesicles 2017, 6, 1369805.
- Khurana, R.; Ranches, G.; Schafferer, S.; Lukasser, M.; Rudnicki, M.; Mayer, G.; Huttenhofer, A. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA 2017, 23, 142–152.
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 2016, 6, 19413.
- Ge, X.; Tang, L.; Wang, Y.; Wang, N.; Zhou, J.; Deng, X.; Zhong, Y.; Li, Q.; Wang, F.; Jiang, G.; et al. The diagnostic value of exosomal miRNAs in human bile of malignant biliary obstructions. Dig. Liver Dis. 2021, 53, 760–765.
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978.
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383.
- Andaloussi, S.E.L.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357.
- Torres, A.G.; Marti, E. Toward an Understanding of Extracellular tRNA Biology. Front. Mol. Biosci. 2021, 8, 662620.
- Schimmel, P. The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018, 19, 45–58.
- Holley, R.W.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquisee, M.; Merrill, S.H.; Penswick, J.R.; Zamir, A. Structure of a Ribonucleic Acid. Science 1965, 147, 1462–1465.
- Novoa, E.M.; Pavon-Eternod, M.; Pan, T.; Ribas de Pouplana, L. A role for tRNA modifications in genome structure and codon usage. Cell 2012, 149, 202–213.
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42.
- Fujishima, K.; Sugahara, J.; Kikuta, K.; Hirano, R.; Sato, A.; Tomita, M.; Kanai, A. Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl. Acad. Sci. USA 2009, 106, 2683–2687.
- Gebetsberger, J.; Polacek, N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013, 10, 1798–1806.
- Tang, S.; Cheng, J.; Yao, Y.; Lou, C.; Wang, L.; Huang, X.; Zhang, Y. Combination of Four Serum Exosomal MiRNAs as Novel Diagnostic Biomarkers for Early-Stage Gastric Cancer. Front. Genet. 2020, 11, 237.
- Cooke, W.R.; Cribbs, A.; Zhang, W.; Kandzija, N.; Motta-Mejia, C.; Dombi, E.; Ri, R.; Cerdeira, A.S.; Redman, C.; Vatish, M. Maternal circulating syncytiotrophoblast-derived extracellular vesicles contain biologically active 5′-tRNA halves. Biochem. Biophys. Res. Commun. 2019, 518, 107–113.
- Yang, Y.; Wang, Y.; Wei, S.; Zhou, C.; Yu, J.; Wang, G.; Wang, W.; Zhao, L. Extracellular vesicles isolated by size-exclusion chromatography present suitability for RNomics analysis in plasma. J. Transl. Med. 2021, 19, 104.
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039.
- Ren, J.; Zhou, Q.; Li, H.; Li, J.; Pang, L.; Su, L.; Gu, Q.; Zhu, Z.; Liu, B. Characterization of exosomal RNAs derived from human gastric cancer cells by deep sequencing. Tumor Biol. 2017, 39, 1–12.
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319.
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995.
- Tosar, J.P.; Rovira, C.; Cayota, A. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol. 2018, 1, 2.
- Amorim, M.G.; Valieris, R.; Drummond, R.D.; Pizzi, M.P.; Freitas, V.M.; Sinigaglia-Coimbra, R.; Calin, G.A.; Pasqualini, R.; Arap, W.; Silva, I.T.; et al. A total transcriptome profiling method for plasma-derived extracellular vesicles: Applications for liquid biopsies. Sci. Rep. 2017, 7, 14395.
- Alexander, R.P.; Kitchen, R.R.; Tosar, J.P.; Roth, M.; Mestdagh, P.; Max, K.E.A.; Rozowsky, J.; Kaczor-Urbanowicz, K.E.; Chang, J.; Balaj, L.; et al. Open Problems in Extracellular RNA Data Analysis: Insights From an ERCC Online Workshop. Front. Genet. 2021, 12, 778416.
- A Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968.
- Kuscu, C.; Kumar, P.; Kiran, M.; Su, Z.; Malik, A.; Dutta, A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 2018, 24, 1093–1105.
- Magee, R.; Rigoutsos, I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res 2020, 48, 9433–9448.
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020, 13, 121.
- Pekarsky, Y.; Balatti, V.; Palamarchuk, A.; Rizzotto, L.; Veneziano, D.; Nigita, G.; Rassenti, L.Z.; Pass, H.I.; Kipps, T.J.; Liu, C.G.; et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 5071–5076.
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473.
- Cheng, W.C.; Liao, T.T.; Lin, C.C.; Yuan, L.E.; Lan, H.Y.; Lin, H.H.; Teng, H.W.; Chang, H.C.; Lin, C.H.; Yang, C.Y.; et al. RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int. J. Cancer 2019, 145, 2209–2224.
- Mao, J.; Liang, Z.; Zhang, B.; Yang, H.; Li, X.; Fu, H.; Zhang, X.; Yan, Y.; Xu, W.; Qian, H. UBR2 Enriched in p53 Deficient Mouse Bone Marrow Mesenchymal Stem Cell-Exosome Promoted Gastric Cancer Progression via Wnt/beta-Catenin Pathway. Stem Cells 2017, 35, 2267–2279.
- Sun, Z.; Wang, L.; Zhou, Y.; Dong, L.; Ma, W.; Lv, L.; Zhang, J.; Wang, X. Glioblastoma Stem Cell-Derived Exosomes Enhance Stemness and Tumorigenicity of Glioma Cells by Transferring Notch1 Protein. Cell. Mol. Neurobiol. 2020, 40, 767–784.
- Baglio, S.R.; Rooijers, K.; Koppers-Lalic, D.; Verweij, F.J.; Perez Lanzon, M.; Zini, N.; Naaijkens, B.; Perut, F.; Niessen, H.W.; Baldini, N.; et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015, 6, 127.
- Vergauwen, G.; Tulkens, J.; Pinheiro, C.; Avila Cobos, F.; Dedeyne, S.; De Scheerder, M.A.; Vandekerckhove, L.; Impens, F.; Miinalainen, I.; Braems, G.; et al. Robust sequential biophysical fractionation of blood plasma to study variations in the biomolecular landscape of systemically circulating extracellular vesicles across clinical conditions. J. Extracell. Vesicles 2021, 10, e12122.
- Kaur, S.; Elkahloun, A.G.; Arakelyan, A.; Young, L.; Myers, T.G.; Otaizo-Carrasquero, F.; Wu, W.; Margolis, L.; Roberts, D.D. CD63, MHC class 1, and CD47 identify subsets of extracellular vesicles containing distinct populations of noncoding RNAs. Sci. Rep. 2018, 8, 2577.
- Gambaro, F.; Calzi, M.L.; Fagundez, P.; Costa, B.; Greif, G.; Mallick, E.; Lyons, S.; Ivanov, P.; Witwer, K.; Cayota, A.; et al. Stable tRNA halves can be sorted into extracellular vesicles and delivered to recipient cells in a concentration-dependent manner. RNA Biol. 2020, 17, 1168–1182.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
682
Revisions:
3 times
(View History)
Update Date:
21 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




