Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zahoor Ahmad Parray | -- | 2878 | 2022-04-19 06:57:33 | | | |
| 2 | Catherine Yang | -5 word(s) | 2873 | 2022-04-19 08:14:03 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Parray, Z.; Islam, A.; , . Protein Structure Fluctuation. Encyclopedia. Available online: https://encyclopedia.pub/entry/21912 (accessed on 07 February 2026).
Parray Z, Islam A, . Protein Structure Fluctuation. Encyclopedia. Available at: https://encyclopedia.pub/entry/21912. Accessed February 07, 2026.
Parray, Zahoor, Asimul Islam, . "Protein Structure Fluctuation" Encyclopedia, https://encyclopedia.pub/entry/21912 (accessed February 07, 2026).
Parray, Z., Islam, A., & , . (2022, April 19). Protein Structure Fluctuation. In Encyclopedia. https://encyclopedia.pub/entry/21912
Parray, Zahoor, et al. "Protein Structure Fluctuation." Encyclopedia. Web. 19 April, 2022.
Copy Citation
Proteins are indispensable to cellular communication and metabolism. The structure on which cells and tissues are developed is deciphered from proteins. To perform functions, proteins fold into a three-dimensional structural design, which is specific and fundamentally determined by their characteristic sequence of amino acids.
protein folding
intermediate states
biological functions
1. Introduction
The regulatory functions of proteins under in vitro complex systems and within the cell are well known. However, proteins that are unfolded or partially folded (intermediates) also play a significant role in different cellular processes and signaling events [1][2][3][4][5]. The role of such intermediates of protein folding has not been discussed in detail until now, and new findings are evolving to provide a fertile ground for considering the molecular mechanisms of biological processes [3][6]. The intermediate state of the protein with a native-like secondary structure but with an unstable or molten tertiary structure can be helpful in understanding pathways of protein folding [7][8]. Such intermediates offer new insights into the role of structural change in proteins within the cell, where transitional states of proteins can be imported and exported more efficiently via membranes than native forms of proteins [5][9]. The translocation of phospholipids between the two monolayers of a lipid bilayer of a cell membrane is carried out by a class of proteins called scramblases. It has been observed by researchers that scramblases are in a fully open state while they assume intermediate states and assist in the transport of ions [10]. These proteins that began as scramblases and were developed into pure ion channels, as a result of mutations, favor the intermediate type [10][11].
2. Intermediate States of Proteins and Their Types
Almost all proteins fold via several partially structured intermediates. To comprehend the structure and structural characteristics of intermediates at the atomic level is often an argumentative content since these are characterized and monitored under an extreme environment of temperature, pH, and chemical denaturants. Besides, chemical modifications, site-directed mutagenesis (or point mutation), and cleavage of the covalent bond of natural proteins are several other routes that often lead to native and/or denatured-like intermediate structures include molten globule (MG) and premolten globule (PMG) states, respectively [12][13][14][15][16].
The molten globule (MG) states are partially unfolded structured forms enfolded with a prominent amount of a secondary structure but a largely chaotic tertiary structure [12][17][18][19][20]. These are compact and native-like structures of the protein considered to be general intermediate states in protein folding [12][21][22]. Because of their similarity to early kinetic intermediate states [18][23], MGs have been proposed as models for transient intermediates in protein folding. The first report of MG state was observed in 1981 [24], while the term MG state was coined in 1983 [23]. Furthermore, the MG states are classified into dry MGs (DMGs) and wet MGs (WMGs). In comparison with the native protein, the former state has slightly extended forms and dry interiors with more conformational flexibility [12][16][25], and the latter possesses hydrated cores with significantly reduced packing in similarity to the folded state [12][25]. Understanding protein folding problems necessitates a comprehensive insight into the characteristics of intermediate species and provides a clear proof of the importance of maintaining proper stoichiometry (as defined by the experimentally observed relative frequencies of amino acids) [26][27]. The following are the common structural features of MGs [17][28]: (i) the presence of a substantial amount of secondary structure (very comparable with that of the protein in native condition) is confirmed by far-UV circular dichroism (CD) and IR spectroscopy, but generally reduced stability of the constitutive hydrogen bonds as represented by proton exchange using 1H NMR [29][30]; (ii) the majority of the particular tertiary structure created by the close packing of side chains is missing as determined by near-UV CD and 2D nuclear magnetic resonance (NMR) [31]; (iii) the protein molecule compactness is with a radius of gyration 10–30% greater than that of the native state [32][33] or a hydrodynamic radius 15–16% greater than that of the native state [23][34]; and (iv) the solvent-exposed loosely packed hydrophobic patches (hydrophobic surface areas) are present due to which it acts sticky [35] and binds to the hydrophobic molecules, such as 8-anilino-1-naphthalenesulfonic acid (ANS) [36] and the Nile red [37].
The premolten globule (PMG) state is less condensed than the MG and native states, but it is far more compacted than the unfolded state (random coil) [38]. Jeng and Englander [38] coined the term PMG in 1991. It is a partially unfolded form of the protein that is believed to be a general protein folding intermediate [18]. PMG states were discovered in several proteins during equilibrium intermediate studies, and are thus considered a fundamental thermodynamic state of the hierarchical protein folding processes [38][39][40][41]. For the past two decades, protein scientists have been intrigued by the PMG, not just because it provides insights into the classic three-stage unfolding process seen in many proteins, but also because it is comparable to the partly folded intermediate temporarily accumulating in the initial stages of folding. Many proteins’ PMG states have been successfully characterized under salt-induced denaturing conditions, such as LiCl and LiClO3 [39][42], by the interaction of many other divalent and trivalent metal ions, including Zn2+ [43], SDS-induced denaturing circumstances, and acidic pH [44][45], and more studies are there where PMG states of different proteins were successfully characterized [38][41][44][46][47][48]. The PMG state is an equilibrium counterpart of the first kinetic folding intermediate formed within a few milliseconds (referred to as the burst-phase intermediate) and accumulates momentarily during refolding from a fully unfolded state [34][45]. The common structural characteristics of PMGs [49][50][51] are: (i) about 50% of the native secondary structure is present, which is revealed from far-UV CD and IR spectroscopy; (ii) no rigid tertiary structure is present as determined by near-UV CD; (iii) compactness (in terms of hydrodynamic volume) is roughly three times greater than that of the N state; and (iv) it shows almost five times weaker ANS binding than for the MG state. It is also widely understood that the protein molecule in the PMG state lacks a globular form, hinting that the PMG is most probably a squeezed, partially structured, and partially disordered conformation of a coil [48][52]. Finally, an all-or-none transition separates the PMG from the MG, which is an intramolecular analog of the first-order step transition [33][39][46][50]. These observations disclosed that both intermediate states (MG and PMG) characterize diverse thermodynamic states of globular proteins. A model has been proposed based on the above knowledge about folded, MG, PMG, and unfolded forms of proteins [16](see Figure 1). Recently, both the computational and spectroscopic approaches were exploited for the successful characterization of two intermediate states (MG and PMG) in myoglobin (Mb) induced by two different concentrations of PEG 4 kDa [53].
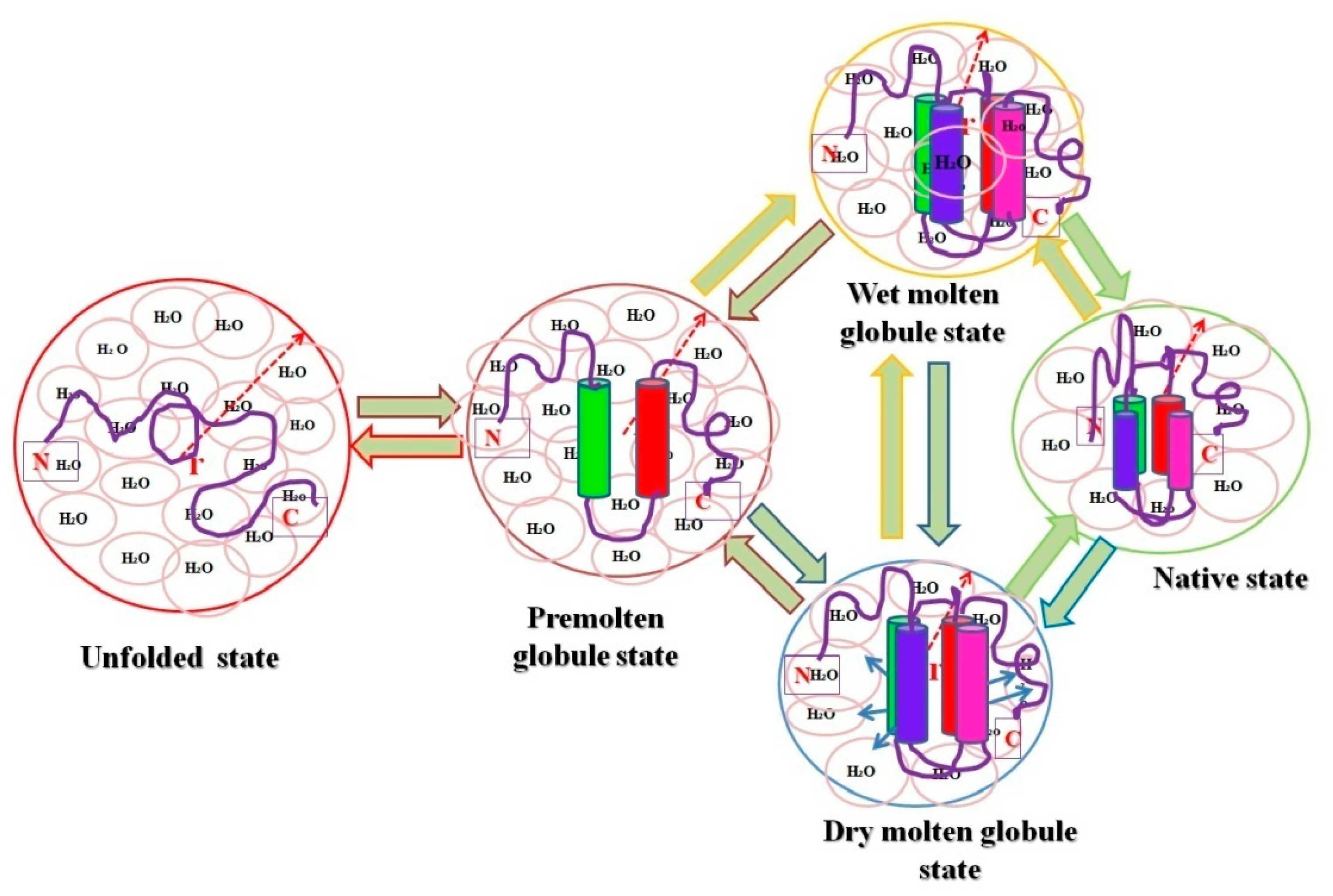
Figure 1. Pictorial representation of relative hydrodynamic volumes of different intermediate states of proteins. The figure shows an ordered secondary structure (cylinder shaped) and water molecules associated with each state (pink circles), and the arrows in dry molten globule (DMG) state represent an increase in the size towards the native state exclusive of water diffusion.
2.1. Intermediate States Characterized under In Vitro Conditions
The intermediate states have been exerting a pull in recent times on scientists who put forth research on protein folding mechanisms to present hints for understanding the classical two-state and/or three-state unfolding methods. The first report on the MG state was observed in 1981, where the heat capacity function in the MG state of apo-α-lactalbumin was examined by a scanning microcalorimeter under physiological pH [24]. Observations of this study showed an enthalpy variation between the MG state and unfolded state (assumed) at neutral pH, which was observed to be almost zero, signifying that the MG state does not show sign any co-operative transition upon heating [24]. Preceding two decades, another third state, measured as a new thermodynamic state of the hierarchical protein folding process called the PMG, has drawn interest among scientists in the protein folding research field because it presents intimations to comprehend the classical three-phase mechanism in unfolding, observed in many proteins [15][38][39][40][41][44][46][47][48]. In vitro experiments have revealed that proteins can be guided to the MG state at acidic pH or high temperatures or in moderate doses of chemical denaturants [54][55]. Many proteins belonging to the structural class of all α or α + β have the majority of MG states characterized and classified in their folding/unfolding routes [56]. Interestingly, only a few studies of proteins found to be fit into all β-sheet categories, which were recognized as an MG state(s) [57][58][59]. Downhill folding mechanisms were intended to exist effectively for proteins with highly optimized native interactions under extremely stable conditions [60][61] or when constructive mutations take place [62]. Their study concluded that at least in metalloproteins, downhill folding can occur under a much greater choice of conditions and can be associated with a variety of other transitions [13]. According to the study, the bacterial zinc finger protein Ros87 has a bipartite folding/unfolding process in which a metal-binding intermediate converts to the native structure via a sensitive barrierless downhill transition. These intermediates were examined using DSC, CD, and NMR in a range of pH, temperature, and ionic strength parameters, showing that the downhill mechanism can be discovered under a considerably broader range of conditions and can be related to a variety of other transitions [13].
2.2. Significance of Intermediary States under In Vivo Conditions
Taking benefits from the protein folding (new wing) (i.e., intermediate states in the cellular conditions) upholds cellular protein homeostasis (proteostasis), which is critical for cell function and development [63][64]. Besides, the folding process—these intermediates assist in many genetic illnesses [8][65][66]. Proteostasis is governed in cellular conditions by networks of protein complexes that include the translation machinery [67][68], proteases [69][70], ubiquitin–proteasome system (UPS) [71], secretory pathways [72][73], autophagic machinery [74], and molecular chaperones [3], which have a significant role in protein homeostasis. To illustrate, a non-native compact type of cyt c is implicated in programmed cell death (induces apoptosis), after which the protein is released from the mitochondrion; non-native forms of the protein are also associated in several of the amyloid-related illnesses [8]. Characterizing the heterogeneity present within the process of folding and unfolding proteins, intermediate states are vital to understanding intermediates and defining their boundaries. The cell intermediate states can be defined as attractors on a potential landscape [1][75][76].
The intermediates not only help to decipher the enormously complex troubles in protein folding, although this also reveals new insight into the importance of structural changes in proteins within cells, whereas protein intermediates can be imported and exported more easily through membranes than native proteins [2][3][77]. The native ⇔ molten globule transition is also considered because the conversion of a protein’s native state to a condensed intermediate structure might occasionally allow it to perform different physiological activities inside the cell [8]. A non-native compact conformation of cyt c, for example, is linked to programmed cell death (apoptosis), whereupon the protein is released from the mitochondrion; non-native forms of the protein are also linked to various amyloid-related diseases [8]. Nuclear genes code for the majority of mitochondrial proteins, which are formed on cytoplasmic ribosomes and transferred into mitochondrial subcompartments [2][78]. To preserve the integrity of protein function in cellular compartments, protein sorting and transport through the lipid membrane of the mitochondrion is desired without intervening with the organelle’s integrity or functions. To understand this to a greater extent, molecular specificity and targeting of mitochondrial preprotein mechanisms and postproteins after import–export via an inner membrane and outer membrane facilitates recognition or identification and is characterized by cellular signaling [2][78]. The presence of these intermediate structures of protein has a significant role in transport via membranes in cellular conditions. The purpose of a set of proteins identified as heat-shock proteins or molecular chaperones located both outside and within the mitochondrion are intimately connected to the unfolding and folding of proteins during transmembrane movement. Investigating the folding of polypeptides in the mitochondrial matrix has provided new and unique findings into general protein folding pathways supported by folding factors [2][78]. Folding and misfolding of proteins in the human membrane help in the resolution of problems related to health and diseases [79]. The new perspective that links membrane protein folding energetics with the degree of complexity of biological systems is recognized via intermediates that play an essential role in the import–export of native protein via membranes and can easily interact with the drug to cure diseases. These advancements in the production of therapeutics and precision medicine are influenced by these intermediate structures in cells [79][80]. From Anfinsen’s experiments [81], which provided how proteins choose their structural elements from denatured conformations and each fraction competes for renaturation to native state [79][82]. The complexity of protein folding makes it difficult to comprehend and even describe the process. Much of this heterogeneity can be described and understood using a statistical approach to the energetics of protein structure (i.e., the energy landscape) [60]. The statistical energy landscape strategy describes why and when particular folding pathways emerge in some proteins, and also how to spot the difference between folding mechanisms that are universal to all sequences and those that are specific to individual sequences. This method also provides fresh quantitative ideas in understanding protein folding thermodynamics and kinetic studies and simulations [60][83].
The ribulose-bis-phosphate carboxylase/oxygenase, Rubisco (an abundant protein on earth), has greater kinetic facets in plants to enhance photosynthesis quality, resulting in species with high nitrogen and water-use efficiencies. This protein improves crop improvement and can provide relief from the CO2 increase caused by anthropogenic activities that lead to global climate changes. Type I Rubisco is a highly conserved hexa-decameric complex found in cyanobacteria, algae, and plants. It consists of eight large subunits with ~50 kDa molecular mass and eight small subunits with ~15 kDa molecular mass. Another kind of bacterial Rubisco (Type II) is a dimer of large subunits that folds and assembles spontaneously in a GroEL-mediated reaction [84][85]. Whether GroEL/GroES was co-overexpressed or not, the expression of Type I Rubisco (from cyanobacteria) in E. coli did not result in the formation of soluble protein, in comparison with the bacterial process. The investigations noticed that in cyanobacteria, the Rubisco operon holds an ORF (open reading frame) for a protein called RbcX. Apart from the genes for the Rubisco subunits, there are genes for the small and large Rubisco subunits (RbcS and RbcL, correspondingly). The existence of the protein (RbcX) was very less renowned before, but researchers have developed methods in E. coli and express Rubisco upon coexpression of RbcX. A Rubisco-specific chaperone could thus be a crucial step in allowing efficient folding of imported Rubiscos in both prokaryotic and plant systems [65][86]. Figure 2 shows the significant role of intermediate assembly in the folding and assembly of L8S8 Rubisco mediated by GroEL/GroES and RbcX. The study showed that folding process includes steps, (i) the substrate bound (primary state) to the chaperonin complex, (ii) structural characterization of intermediate states kinetically trapped and accumulated throughout the folding route, and (iii) kinetic measurements during the process (unfolded ⇔ intermediate ⇔ native state conversion) [84].
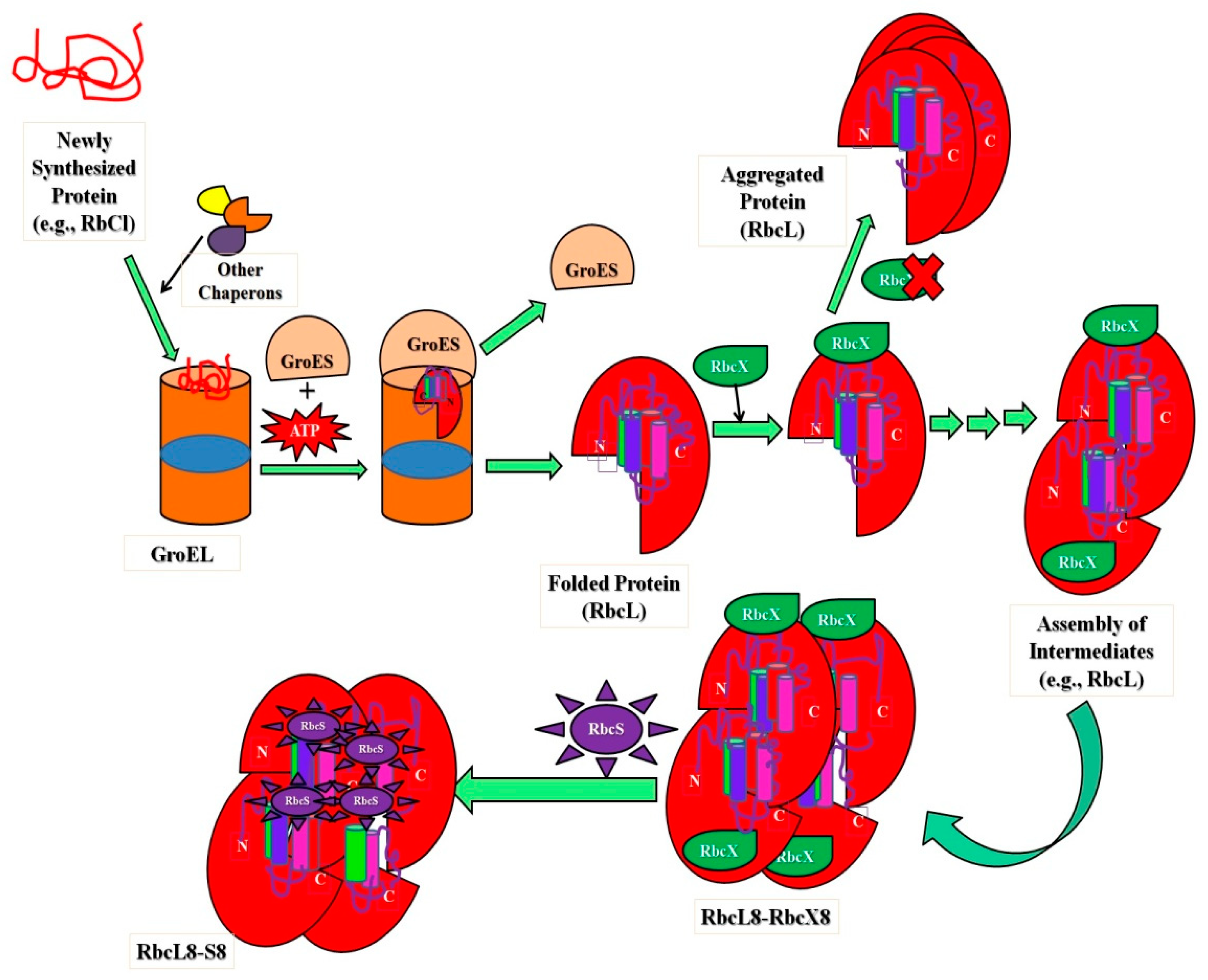
Figure 2. Schematic representation showing the importance of intermediates in folding and assembly of L8S8 Rubisco (cyanobacterial) mediated by GroEL/GroES and RbcX.
In addition, the researchers observed that folding intermediates provide approaches to differences in immunoglobulin amyloidogenicity and thus can shape the folding landscape positively to favor either folding or misfolding [3][71][87][88]. The researchers used an antibody domain’s intrinsically slow folding process to define its essential folding intermediate [87]. They were able to trap the intermediate in equilibrium and identify it at atomic resolution using a single-point mutation. It is also worth noting that intermediate has the simple β-barrel topology; however, a few strands were observed to be distorted [87]. Unexpectedly, the presence of two short-strand-connecting helices in the constant region of antibody domains suggests that a native structure is fully present in the intermediate, which was then used as a framework for subsequent strands [87]. Transplanting these conserved stands of helices into β 2-microglobulin (homologous member of the same superfamily) considerably showed a reduction in its amyloidogenicity [87]. As a result, a high level of local structuring intermediates through protein folding can have a considerable effect on the folding landscape which favored vigorous folding against negative misfolding. In addition, throughout evolution, the small differences acquired amid members of the identical protein superfamily can evade pathogenic misfolding reaction and identical protein topology conservation [87].
As it is a known fact that transferable agents called prions cause spongiform encephalopathies (TSEs) in animals as well as humans. They are made up of PrPSc, the infectious isomer of PrPC, and the cellular prion protein [89]. The conversion and propensity of the protein commence alternative folds, which are liable for the species-specific transmission of the disease. Kachel et al. defined and confirmed the structural stages of the human prion protein (hu PrP) [89] by using a hydrostatic pressure (up to 200 MPa) and two-dimensional NMR spectroscopy in combination. They recognized folding intermediates that were stabilized by pressure of the human prion protein. They observed that the β1/α1-loopand the solvent-exposed side of α3 are the strongest regions reflecting the transition to the intermediate states [89]. Their findings showed that the loop between β-strand 1 and α-helix 1 (residues 139–141) was the most pressure-sensitive region (intermediate I1), and may be the first gateway for the infectious moiety to transform the cellular protein [89].
Therefore, folding intermediates are essential in determining protein folding parameters, understanding protein folding mechanisms, conservation of protein topology, cellular transport regulation, structural maintenance, and avoidance of protein misfolding. These elements are also better for understanding the biologically significant mechanism of conformational changes, such as structural distributions, harmonic vibrations, and structural fluctuations [83][90]. Therefore, it may not be wrong to say that intermediates are vibrant and vigorous elements of cellular architecture.
References
- Parray, Z.A.; Ahmad, F.; Alajmi, M.F.; Hussain, A.; Hassan, M.I.; Islam, A. Interaction of polyethylene glycol with cytochrome c investigated via in vitro and in silico approaches. Sci. Rep. 2021, 11, 6475.
- Alberts, B.; Johnson, A.; Lewis, J. The Transport of Proteins into Mitochondria and Chloroplasts. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002.
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354.
- Banks, A.; Qin, S.; Weiss, K.L.; Stanley, C.B.; Zhou, H.X. Intrinsically Disordered Protein Exhibits Both Compaction and Expansion under Macromolecular Crowding. Biophys. J. 2018, 114, 1067–1079.
- Tsytlonok, M.; Itzhaki, L.S. The how’s and why’s of protein folding intermediates. Arch. Biochem. Biophys. 2013, 531, 14–23.
- Dyson, H.J.; Wright, P.E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004, 104, 3607–3622.
- Veprintsev, D.B.; Permyakov, S.E.; Permyakov, E.A.; Rogov, V.V.; Cawthern, K.M.; Berliner, L.J. Cooperative thermal transitions of bovine and human apo-alpha-lactalbumins: Evidence for a new intermediate state. FEBS Lett. 1997, 412, 625–628.
- Santucci, R.; Polticelli, F.; Sinibaldi, F.; Fiorucci, L. Chapter 14—Role of Intermediate States in Protein Folding and Misfolding. In Recent Advances in Medicinal Chemistry; Atta ur, R., Choudhary, M.I., Perry, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 433–455.
- Boesch, C.; Bundi, A.; Oppliger, M.; Wuthrich, K. 1H nuclear-magnetic-resonance studies of the molecular conformation of monomeric glucagon in aqueous solution. Eur. J. Biochem. 1978, 91, 209–214.
- Daily, S. How a Membrane Protein Can Move Both Lipids and Ions. Science Daily, University of Groningen. 2019. Available online: https://www.sciencedaily.com/releases/2019/03/190312075928.htm (accessed on 20 February 2022).
- Alvadia, C.; Lim, N.K.; Clerico Mosina, V.; Oostergetel, G.T.; Dutzler, R.; Paulino, C. Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. Elife 2019, 8, e44365.
- Bhattacharyya, S.; Varadarajan, R. Packing in molten globules and native states. Curr. Opin. Struct. Biol. 2013, 23, 11–21.
- Grazioso, R.; García-Viñuales, S.; D’Abrosca, G.; Baglivo, I.; Pedone, P.V.; Milardi, D.; Fattorusso, R.; Isernia, C.; Russo, L.; Malgieri, G. The change of conditions does not affect Ros87 downhill folding mechanism. Sci. Rep. 2020, 10, 21067.
- Khan, S.H.; Islam, A.; Hassan, M.I.; Sharma, S.; Singh, T.P.; Ahmad, F. Effect of conservative mutations (L94V and L94I) on the structure and stability of horse cytochrome c. Arch. Biochem. Biophys. 2017, 633, 40–49.
- Khan, M.K.; Rahaman, H.; Ahmad, F. Conformation and thermodynamic stability of pre-molten and molten globule states of mammalian cytochromes-c. Metallomics 2011, 3, 327–338.
- Naiyer, A.; Hassan, M.I.; Islam, A.; Sundd, M.; Ahmad, F. Structural characterization of MG and pre-MG states of proteins by MD simulations, NMR, and other techniques. J. Biomol. Struct. Dyn. 2015, 33, 2267–2284.
- Parray, Z.A.; Shahid, S.; Ahmad, F.; Hassan, M.I.; Islam, A. Characterization of intermediate state of myoglobin in the presence of PEG 10 under physiological conditions. Int. J. Biol. Macromol. 2017, 99, 241–248.
- Arai, M.; Kuwajima, K. Role of the molten globule state in protein folding. Adv. Protein Chem. 2000, 53, 209–282.
- Kim, P.S.; Baldwin, R.L. Intermediates in the folding reactions of small proteins. Annu. Rev. Biochem. 1990, 59, 631–660.
- Baldwin, R.L. Experimental studies of pathways of protein folding. Ciba Found. Symp. 1991, 161, 190–201.
- Ptitsyn, O.B.; Uversky, V.N. The molten globule is a third thermodynamical state of protein molecules. FEBS Lett. 1994, 341, 15–18.
- Kuwajima, K.; Nitta, K.; Yoneyama, M.; Sugai, S. Three-state denaturation of alpha-lactalbumin by guanidine hydrochloride. J. Mol. Biol. 1976, 106, 359–373.
- Ohgushi, M.; Wada, A. ‘Molten-globule state’: A compact form of globular proteins with mobile side-chains. FEBS Lett. 1983, 164, 21–24.
- Yutani, K.; Ogasahara, K.; Kuwajima, K. Absence of the thermal transition in apo-α-lactalbumin in the molten globule state: A study by differential scanning microcalorimetry. J. Mol. Biol. 1992, 228, 347–350.
- Baldwin, R.L.; Rose, G.D. Molten globules, entropy-driven conformational change and protein folding. Curr. Opin. Struct. Biol. 2013, 23, 4–10.
- Mezei, M. Discriminatory power of stoichiometry-driven protein folding? J. Biomol. Struct. Dyn. 2011, 28, 625–626.
- Nagaraj, R. Is protein folding still a challenge? J. Biomol. Struct. Dyn. 2011, 28, 639–640.
- Kuwajima, K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins 1989, 6, 87–103.
- Hughson, F.M.; Wright, P.E.; Baldwin, R.L. Structural characterization of a partly folded apomyoglobin intermediate. Science 1990, 249, 1544–1548.
- Baum, J.; Dobson, C.M.; Evans, P.A.; Hanley, C. Characterization of a partly folded protein by NMR methods: Studies on the molten globule state of guinea pig alpha-lactalbumin. Biochemistry 1989, 28, 7–13.
- Jeng, M.F.; Englander, S.W.; Elove, G.A.; Roder, H.; Wand, A.J. Structural description of acid-denatured cytochrome c by hydrogen exchange and 2D NMR. Biochemistry 1990, 29, 10433–10437.
- Hamada, D.; Kuroda, Y.; Kataoka, M.; Aimoto, S.; Yoshimura, T.; Goto, Y. Role of heme axial ligands in the conformational stability of the native and molten globule states of horse cytochrome c. J. Mol. Biol. 1996, 256, 172–186.
- Uversky, V.N.; Ptitsyn, O.B. “Partly Folded” State, a New Equilibrium State of Protein Molecules: Four-State Guanidinium Chloride-Induced Unfolding of. beta.-Lactamase at Low Temperature. Biochemistry 1994, 33, 2782–2791.
- Uversky, V.N.; Li, J.; Fink, A.L. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson’s disease and heavy metal exposure. J. Biol. Chem. 2001, 276, 44284–44296.
- Ptitsyn, O.B. Protein evolution and protein folding: Non-functional conserved residues and their probable role. In Proceedings of the Pacific Symposium on Biocomputing, PSB 1999, The Big Island, HI, USA, 4–9 January 1999; pp. 494–504.
- Semisotnov, G.V.; Rodionova, N.A.; Razgulyaev, O.I.; Uversky, V.N.; Gripas, A.F.; Gilmanshin, R.I. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 1991, 31, 119–128.
- Sackett, D.L.; Wolff, J. Nile red as a polarity-sensitive fluorescent probe of hydrophobic protein surfaces. Anal. Biochem. 1987, 167, 228–234.
- Jeng, M.F.; Englander, S.W. Stable submolecular folding units in a non-compact form of cytochrome c. J. Mol. Biol. 1991, 221, 1045–1061.
- Alam Khan, M.K.; Das, U.; Rahaman, M.H.; Hassan, M.I.; Srinivasan, A.; Singh, T.P.; Ahmad, F. A single mutation induces molten globule formation and a drastic destabilization of wild-type cytochrome c at pH 6.0. J. Biol. Inorg. Chem. 2009, 14, 751–760.
- Haque, M.A.; Ubaid-Ullah, S.; Zaidi, S.; Hassan, M.I.; Islam, A.; Batra, J.K.; Ahmad, F. Characterization of pre-molten globule state of yeast iso-1-cytochrome c and its deletants at pH 6.0 and 25 °C. Int. J. Biol. Macromol. 2015, 72, 1406–1418.
- Tcherkasskaya, O.; Uversky, V.N. Polymeric aspects of protein folding: A brief overview. Protein Pept. Lett. 2003, 10, 239–245.
- Alam Khan, M.K.; Rahaman, M.H.; Hassan, M.I.; Singh, T.P.; Moosavi-Movahedi, A.A.; Ahmad, F. Conformational and thermodynamic characterization of the premolten globule state occurring during unfolding of the molten globule state of cytochrome c. J. Biol. Inorg. Chem. 2010, 15, 1319–1329.
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J. Biol. Chem. 2001, 276, 10737–10744.
- Kumari, N.; Dubey, V.; Jagannadham, M. SDS Induced Refolding of Pre-molten Globule State of Cryptolepain: Differences in Chemical and Temperature-Induced Equilibrium Unfolding of the Protein in SDS-Induced State. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2013, 83, 71–80.
- Uversky, V.N. Size-exclusion chromatography in structural analysis of intrinsically disordered proteins. Methods Mol. Biol. 2012, 896, 179–194.
- Uversky, V.N.; Ptitsyn, O.B. Further evidence on the equilibrium “pre-molten globule state”: Four-state guanidinium chloride-induced unfolding of carbonic anhydrase B at low temperature. J. Mol. Biol. 1996, 255, 215–228.
- Parray, Z.A.; Ahamad, S.; Ahmad, F.; Hassan, M.I.; Islam, A. First evidence of formation of pre-molten globule state in myoglobin: A macromolecular crowding approach towards protein folding in vivo. Int. J. Biol. Macromol. 2019, 126, 1288–1294.
- Uversky, V.N.; Fink, A.L. Do protein molecules have a native-like topology in the pre-molten globule state? Biochemistry 1999, 64, 552–555.
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756.
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12.
- Uversky, V.N.; Narizhneva, N.V.; Kirschstein, S.O.; Winter, S.; Lober, G. Conformational transitions provoked by organic solvents in beta-lactoglobulin: Can a molten globule like intermediate be induced by the decrease in dielectric constant? Fold. Des. 1997, 2, 163–172.
- Uversky, V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: Which way to go? Cell. Mol. Life Sci. 2003, 60, 1852–1871.
- Parray, Z.A.; Naqvi, A.A.T.; Ahmad, F.; Hassan, M.I.; Islam, A. Characterization of different intermediate states in myoglobin induced by polyethylene glycol: A process of spontaneous molecular self-organization foresees the energy landscape theory via in vitro and in silico approaches. J. Mol. Liq. 2021, 342, 117502.
- Ptitsyn, O.B.; Creighton, T.E. Protein Folding; W.H. Freeman and Co.: New York, NY, USA, 1992; pp. 243–300.
- Samuel, D.; Kumar, T.; Srimathi, T.; Hsieh, H.c.; Yu, C. Identification and Characterization of an Equilibrium Intermediate in the Unfolding Pathway of an All β-Barrel Protein. J. Biol. Chem. 2000, 275, 34968–34975.
- Dobson, C.M.; Karplus, M. The fundamentals of protein folding: Bringing together theory and experiment. Curr. Opin. Struct. Biol. 1999, 9, 92–101.
- Liu, Z.P.; Rizo, J.; Gierasch, L.M. Equilibrium folding studies of cellular retinoic acid binding protein, a predominantly beta-sheet protein. Biochemistry 1994, 33, 134–142.
- Sivaraman, T.; Kumar, T.K.; Jayaraman, G.; Han, C.C.; Yu, C. Characterization of a partially structured state in an all-beta-sheet protein. Biochem. J. 1997, 321 Pt 2, 457–464.
- Ferguson, N.; Capaldi, A.P.; James, R.; Kleanthous, C.; Radford, S.E. Rapid folding with and without populated intermediates in the homologous four-helix proteins Im7 and Im9. J. Mol. Biol. 1999, 286, 1597–1608.
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins 1995, 21, 167–195.
- Gruebele, M. Downhill protein folding: Evolution meets physics. Comptes Rendus Biol. 2005, 328, 701–712.
- Johnson, C.M.; Sharpe, T.D. Protein Folding, Energy Landscapes and Downhill Protein Folding Scenarios. In Encyclopedia of Biophysics; Roberts, G., Watts, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–19.
- Powers, M.V.; Workman, P. Inhibitors of the heat shock response: Biology and pharmacology. FEBS Lett. 2007, 581, 3758–3769.
- Yu, A.; Fox, S.G.; Cavallini, A.; Kerridge, C.; O’Neill, M.J.; Wolak, J.; Bose, S.; Morimoto, R.I. Tau protein aggregates inhibit the protein-folding and vesicular trafficking arms of the cellular proteostasis network. J. Biol. Chem. 2019, 294, 7917–7930.
- Saschenbrecker, S.; Bracher, A.; Rao, K.V.; Rao, B.V.; Hartl, F.U.; Hayer-Hartl, M. Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 2007, 129, 1189–1200.
- Shahnaj, S.; Potshangbam, A.M.; Chowhan, R.K.; Parray, Z.A.; Kakchingtabam, P.; Kumari, A.; Islam, A.; Khan, A.; Singh, L.R.; Rahaman, H. The anti-oxidant enzyme, Prdx6 might have cis-acting regulatory sequence(s). Int. J. Biol. Macromol. 2020, 149, 1139–1150.
- Steffen, K.K.; Dillin, A. A Ribosomal Perspective on Proteostasis and Aging. Cell Metab. 2016, 23, 1004–1012.
- Anisimova, A.S.; Alexandrov, A.I.; Makarova, N.E.; Gladyshev, V.N.; Dmitriev, S.E. Protein synthesis and quality control in aging. Aging 2018, 10, 4269–4288.
- Wilson, C.H.; Shalini, S.; Filipovska, A.; Richman, T.R.; Davies, S.; Martin, S.D.; McGee, S.L.; Puccini, J.; Nikolic, A.; Dorstyn, L.; et al. Age-related proteostasis and metabolic alterations in Caspase-2-deficient mice. Cell Death Dis. 2015, 6, e1615.
- Klein, T.; Eckhard, U.; Dufour, A.; Solis, N.; Overall, C.M. Proteolytic Cleavage-Mechanisms, Function, and ”Omic” Approaches for a Near-Ubiquitous Posttranslational Modification. Chem. Rev. 2018, 118, 1137–1168.
- Bett, J.S. Proteostasis regulation by the ubiquitin system. Essays Biochem. 2016, 60, 143–151.
- Wang, Y.; Lomakin, A.; Kanai, S.; Alex, R.; Benedek, G.B. Liquid–Liquid Phase Separation in Oligomeric Peptide Solutions. Langmuir 2017, 33, 7715–7721.
- Remondelli, P.; Renna, M. The Endoplasmic Reticulum Unfolded Protein Response in Neurodegenerative Disorders and Its Potential Therapeutic Significance. Front. Mol. Neurosci. 2017, 10, 187.
- Carra, S.; Brunsting, J.F.; Lambert, H.; Landry, J.; Kampinga, H.H. HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2α phosphorylation. J. Biol. Chem. 2009, 284, 5523–5532.
- Parray, Z.A.; Hassan, M.I.; Ahmad, F.; Islam, A. Amphiphilic nature of polyethylene glycols and their role in medical research. Polym. Test. 2020, 82, 106316.
- Harada, R.; Inagaki, Y.; Shigeta, Y. Protein Folding and Evolution. In Reference Module in Materials Science and Materials Engineering; Elsevier: Amsterdam, The Netherlands, 2016.
- Amara, J.F.; Cheng, S.H.; Smith, A.E. Intracellular protein trafficking defects in human disease. Trends Cell. Biol. 1992, 2, 145–149.
- Neupert, W. Transport of proteins across mitochondrial membranes. Clin. Investig. 1994, 72, 251–261.
- Marinko, J.T.; Huang, H.; Penn, W.D.; Capra, J.A.; Schlebach, J.P.; Sanders, C.R. Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis. Chem. Rev. 2019, 119, 5537–5606.
- Bhat, M.Y.; Dar, T.A.; Singh, L.R. Casein Proteins: Structural and Functional Aspects, Milk Proteins—From Structure to Biological Properties and Health Aspects; IntechOpen: London, UK, 2016.
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230.
- Rose, G.D.; Fleming, P.J.; Banavar, J.R.; Maritan, A. A backbone-based theory of protein folding. Proc. Natl. Acad. Sci. USA 2006, 103, 16623–16633.
- Parak, F.G. Proteins in action: The physics of structural fluctuations and conformational changes. Curr. Opin. Struct. Biol. 2003, 13, 552–557.
- Lin, Z.; Rye, H.S. GroEL-mediated protein folding: Making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 211–239.
- Brinker, A.; Pfeifer, G.; Kerner, M.J.; Naylor, D.J.; Hartl, F.U.; Hayer-Hartl, M. Dual Function of Protein Confinement in Chaperonin-Assisted Protein Folding. Cell 2001, 107, 223–233.
- Hayer-Hartl, M. From chaperonins to Rubisco assembly and metabolic repair. Protein Sci. 2017, 26, 2324–2333.
- Feige, M.J.; Groscurth, S.; Marcinowski, M.; Yew, Z.T.; Truffault, V.; Paci, E.; Kessler, H.; Buchner, J. The structure of a folding intermediate provides insight into differences in immunoglobulin amyloidogenicity. Proc. Natl. Acad. Sci. USA 2008, 105, 13373–13378.
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Intrinsically disordered proteins in crowded milieu: When chaos prevails within the cellular gumbo. Cell. Mol. Life Sci. CMLS 2018, 75, 3907–3929.
- Kachel, N.; Kremer, W.; Zahn, R.; Kalbitzer, H.R. Observation of intermediate states of the human prion protein by high pressure NMR spectroscopy. BMC Struct. Biol. 2006, 6, 16.
- Gräter, F.; Grubmüller, H. Fluctuations of primary ubiquitin folding intermediates in a force clamp. J. Struct. Biol. 2007, 157, 557–569.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Entry Collection:
Peptides for Health Benefits
Revisions:
2 times
(View History)
Update Date:
19 Apr 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




