
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | david e. scheim | -- | 2965 | 2022-03-31 04:33:48 | | | |
| 2 | Vivi Li | -86 word(s) | 2879 | 2022-03-31 05:43:01 | | |
Video Upload Options
Although COVID-19 typically gains infectious penetration in the respiratory epithelium, vascular damage is frequently observed in lungs and other organ systems of COVID-19 patients, with morbidities such as intravascular clotting, microvascular occlusion and peripheral ischemia. The arrangement and chemical composition of the glycans at the 22 N-glycosylation sites of SARS-CoV-2 spike protein and those at the sialoglycoprotein coating of RBCs allow exploration of specifics as to how virally induced RBC clumping may form. The in vitro and clinical testing of these possibilities can be sharpened by the incorporation of an existing anti-COVID-19 therapeutic that has been found in silico to competitively bind to multiple glycans on SARS-CoV-2 spike protein.
1. Introduction
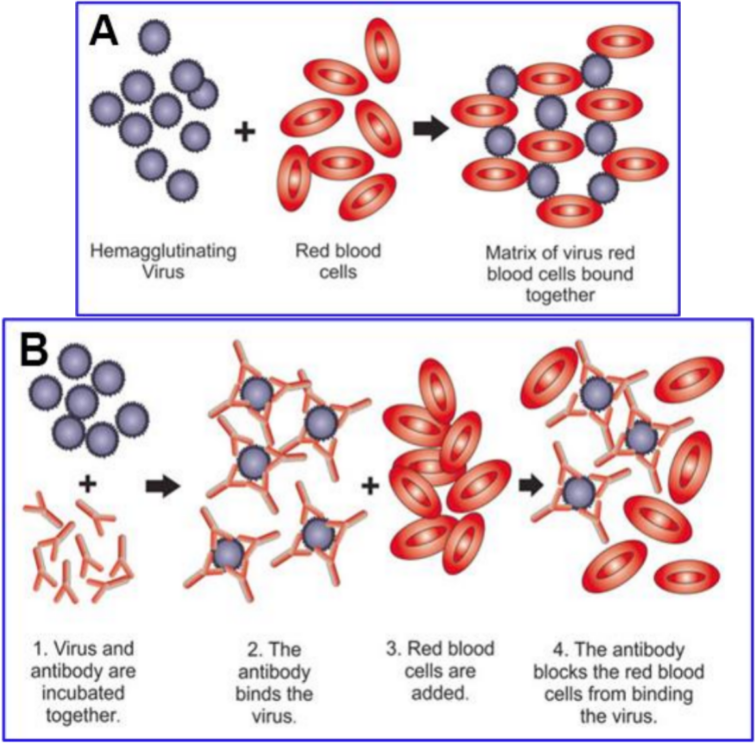
1.1. Binding of Viruses to SA and Host Decoy Defense
1.2. Cleavage of Viral–SA Binding via Hemagglutinin Esterase (HE), an Enzyme Expressed by the Common Cold Betacoronavirus Strains but Not by SARS-CoV, SARS-CoV-2 and MERS
1.3. Multivalent Binding
2. Properties of SARS-CoV-2 Related to Its Array of Spike Glycoprotein Glycans
2.1. SARS-CoV-2 Binds to SA and CD147
2.2. SARS-CoV-2 Attaches to RBCs, Other Blood Cells and Endothelial Cells
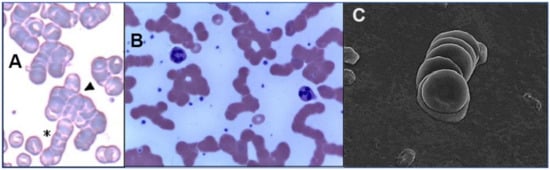
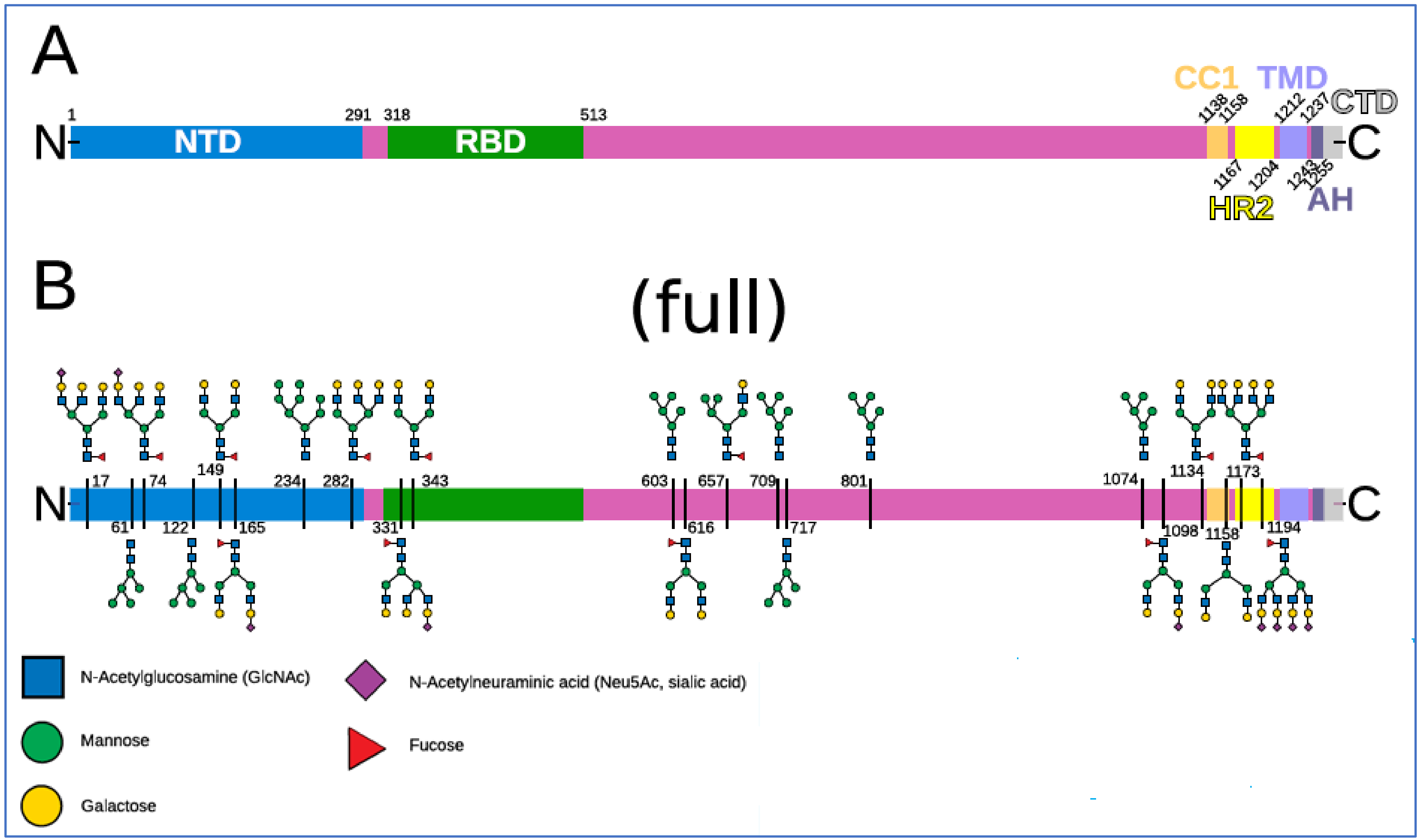
2.3. The Glycan Distribution and Composition at the 22 N-Glycosylation Sites of SARS-CoV-2 Spike Protein
References
- Valyaeva, A.A.; Zharikova, A.A.; Kasianov, A.S.; Vassetzky, Y.S.; Sheval, E.V. Expression of SARS-CoV-2 entry factors in lung epithelial stem cells and its potential implications for COVID-19. Sci. Rep. 2020, 10, 17772.
- Li, H.; Liu, Z.; Ge, J. Scientific research progress of COVID-19/SARS-CoV-2 in the first five months. J. Cell. Mol. Med. 2020, 24, 6558–6570.
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062.
- Couzin-Frankel, J. The mystery of the pandemic’s ‘happy hypoxia’. Science 2020, 368, 455–456.
- Rapkiewicz, A.V.; Mai, X.; Carsons, S.E.; Pittaluga, S.; Kleiner, D.E.; Berger, J.S.; Thomas, S.; Adler, N.M.; Charytan, D.M.; Gasmi, B.; et al. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: A case series. EClinicalMedicine 2020, 24, 100434.
- Negri, E.M.; Piloto, B.M.; Morinaga, L.K.; Jardim, C.V.P.; Lamy, S.A.E.-D.; Ferreira, M.A.; D’Amico, E.A.; Deheinzelin, D. Heparin Therapy Improving Hypoxia in COVID-19 Patients—A Case Series. Front. Physiol. 2020, 11, 1341.
- Lodigiani, C.; Lapichino, G.; Carenzo, L.; Cecconi, M.; Ferrazzi, P.; Sebastian, T.; Kucher, N.; Studt, J.D.; Sacco, C.; Alexia, B.; et al. Venous and arterial thromboembolic complications in COVID-19 patients admitted to an academic hospital in Milan, Italy. Thromb Res. 2020, 191, 9–14.
- Price, L.C.; McCabe, C.; Garfield, B.; Wort, S.J. Thrombosis and COVID-19 pneumonia: The clot thickens! Eur. Respir. J. 2020, 56, 2001608.
- Helms, J.; Tacquard, C.; Severac, F.; Leonard-Lorant, I.; Ohana, M.; Delabranche, X.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Fagot Gandet, F.; et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: A multicenter prospective cohort study. Intensive Care Med. 2020, 46, 1089–1098.
- Huertas, A.; Montani, D.; Savale, L.; Pichon, J.; Tu, L.; Parent, F.; Guignabert, C.; Humbert, M. Endothelial cell dysfunction: A major player in SARS-CoV-2 infection (COVID-19)? Eur. Respir. J. 2020, 56.
- Sardu, C.; Gambardella, J.; Morelli, M.B.; Wang, X.; Marfella, R.; Santulli, G. Hypertension, thrombosis, kidney failure, and diabetes: Is COVID-19 an endothelial disease? A comprehensive evaluation of clinical and basic evidence. J. Clin. Med. 2020, 9, 1417.
- Jung, F.; Krüger-Genge, A.; Franke, R.P.; Hufert, F.; Küpper, J.H. COVID-19 and the endothelium. Clin. Hemorheol. Microcirc. 2020, 75, 7–11.
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032.
- Bikdeli, B.; Madhavan, M.V.; Jimenez, D.; Chuich, T.; Dreyfus, I.; Driggin, E.; Nigoghossian, C.; Ageno, W.; Madjid, M.; Guo, Y.; et al. COVID-19 and thrombotic or thromboembolic disease: Implications for prevention, antithrombotic therapy, and follow-up: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2020, 75, 2950–2973.
- Libby, P.; Lüscher, T. COVID-19 is, in the end, an endothelial disease. Eur. Heart J. 2020, 41, 3038–3044.
- Becker, R.C. COVID-19 update: Covid-19-associated coagulopathy. J. Thromb Thrombolysis 2020, 50, 54–67.
- Mondal, R.; Lahiri, D.; Deb, S.; Bandyopadhyay, D.; Shome, G.; Sarkar, S.; Paria, S.R.; Thakurta, T.G.; Singla, P.; Biswas, S.C. COVID-19: Are we dealing with a multisystem vasculopathy in disguise of a viral infection? J. Thromb Thrombolysis 2020, 50, 567–579.
- Magro, C.; Mulvey, J.J.; Berlin, D.; Nuovo, G.; Salvatore, S.; Harp, J.; Baxter-Stoltzfus, A.; Laurence, J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl. Res. 2020, 220, 1–13.
- Menter, T.; Haslbauer, J.D.; Nienhold, R.; Savic, S.; Hopfer, H.; Deigendesch, N.; Frank, S.; Turek, D.; Willi, N.; Pargger, H.; et al. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020, 77, 198–209.
- Gattinoni, L.; Coppola, S.; Cressoni, M.; Busana, M.; Rossi, S.; Chiumello, D. COVID-19 does not lead to a “typical” acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2020, 201, 1299–1300.
- Marini, J.J.; Gattinoni, L. Management of COVID-19 respiratory distress. JAMA 2020, 323, 2329–2330.
- Scheim, D.E. From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate, then Clot Blood Cells in Pulmonary and Systemic Microvasculature. Available online: http://ssrn.com/abstract=3706347 (accessed on 24 November 2021).
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.; et al. Angiotensin-converting enzyme-2 (ACE2), SARS-CoV-2 and pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 25, 228–248.
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904.e899.
- Ströh, L.J.; Stehle, T. Glycan engagement by viruses: Receptor switches and specificity. Annu Rev. Virol 2014, 1, 285–306.
- Milanetti, E.; Miotto, M.; Di Rienzo, L.; Nagaraj, M.; Monti, M.; Golbek, T.W.; Gosti, G.; Roeters, S.J.; Weidner, T.; Otzen, D.E.; et al. In-Silico Evidence for a Two Receptor Based Strategy of SARS-CoV-2. Front Mol Biosci 2021, 8, 690655.
- Morniroli, D.; Giannì, M.L.; Consales, A.; Pietrasanta, C.; Mosca, F. Human sialome and coronavirus disease-2019 (COVID-19) pandemic: An understated correlation? Front. Immunol. 2020, 11, 1480.
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 55, 105960.
- Roe, K. High COVID-19 virus replication rates, the creation of antigen–antibody immune complexes and indirect haemagglutination resulting in thrombosis. Transbound. Emerg. Dis. 2020, 67, 1418–1421.
- Koehler, M.; Delguste, M.; Sieben, C.; Gillet, L.; Alsteens, D. Initial step of virus entry: Virion binding to cell-surface glycans. Annu. Rev. Virol. 2020, 7, 143–165.
- Baum, J.; Ward, R.H.; Conway, D.J. Natural selection on the erythrocyte surface. Mol. Biol. Evol. 2002, 19, 223–229.
- Storry, J.R. Review: The function of blood group-specific RBC membrane components. Immunohematology 2004, 20, 206–216.
- Levine, S.; Levine, M.; Sharp, K.A.; Brooks, D.E. Theory of the electrokinetic behavior of human erythrocytes. Biophys. J. 1983, 42, 127–135.
- Bai, Y.; Huang, W.; Ma, L.T.; Jiang, J.L.; Chen, Z.N. Importance of N-glycosylation on CD147 for its biological functions. Int. J. Mol. Sci. 2014, 15, 6356–6377.
- Modrof, J.; Kerschbaum, A.; Farcet, M.R.; Niemeyer, D.; Corman, V.M.; Kreil, T.R. SARS-CoV-2 and the safety margins of cell-based biological medicinal products. Biologicals 2020, 68, 122–124.
- Lam, L.M.; Murphy, S.J.; Kuri-Cervantes, L.; Weisman, A.R.; Ittner, C.A.G.; Reilly, J.P.; Pampena, M.B.; Betts, M.R.; Wherry, E.J.; Song, W.-C.; et al. Erythrocytes reveal complement activation in patients with COVID-19. medRxiv 2020, 05.20.20104398.
- Howe, C.; Lee, L.T. Virus-erythrocyte interactions. In Advances in Virus Research; Smith, K.M., Lauffer, M.A., Bang, F.B., Eds.; Academic Press: New York, NY, USA, 1972; Volume 17, pp. 1–50.
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic acid receptors of viruses. In SialoGlyco Chemistry and Biology II: Tools and Techniques to Identify and Capture Sialoglycans; Gerardy-Schahn, R., Delannoy, P., von Itzstein, M., Eds.; Springer International Publishing: New York, NY, USA, 2015; pp. 1–28.
- Kapikian, A.Z.; James, H.D., Jr.; Kelly, S.J.; King, L.M.; Vaughn, A.L.; Chanock, R.M. Hemadsorption by coronavirus strain OC43. Proc. Soc. Exp. Biol. Med. 1972, 139, 179–186.
- Agafonov, A.P.; Gus’kov, A.A.; Ternovoi, V.A.; Ryabchikova, E.I.; Durymanov, A.G.; Vinogradov, I.V.; Maksimov, N.L.; Ignat’ev, G.M.; Nechaeva, E.A.; Netesov, S.V. Primary characterization of SARS coronavirus strain Frankfurt 1. Dokl. Biol. Sci. 2004, 394, 58–60.
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529.
- Storz, J.; Zhang, X.M.; Rott, R. Comparison of hemagglutinating, receptor-destroying, and acetylesterase activities of avirulent and virulent bovine coronavirus strains. Arch. Virol. 1992, 125, 193–204.
- Brian, D.A.; Hogue, B.G.; Kienzle, T.E. The coronavirus hemagglutinin esterase glycoprotein. In The Coronaviridae. The Viruses; Siddell, S.G., Ed.; Springer: Boston, MA, USA, 1995.
- Qing, E.; Hantak, M.; Perlman, S.; Gallagher, T. Distinct Roles for sialoside and protein receptors in coronavirus infection. mBio 2020, 11, e02764–e02819.
- Schultze, B.; Cavanagh, D.; Herrler, G. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid-containing receptors on erythrocytes. Virology 1992, 189, 792–794.
- Schultze, B.; Enjuanes, L.; Herrler, G. Analysis of the sialic acid-binding activity of transmissible gastroenteritis virus. In Corona- and Related Viruses: Current Concepts in Molecular Biology and Pathogenesis; Talbot, P.J., Levy, G.A., Eds.; Springer: Boston, MA, USA, 1995; pp. 367–370.
- Hulswit, R.J.G.; de Haan, C.A.M.; Bosch, B.J. Chapter two—Coronavirus spike protein and tropism changes. In Advances in Virus Research; Ziebuhr, J., Ed.; Academic Press: New York, NY, USA, 2016; Volume 96, pp. 29–57.
- Li, W.; Hulswit, R.J.G.; Widjaja, I.; Raj, V.S.; McBride, R.; Peng, W.; Widagdo, W.; Tortorici, M.A.; van Dieren, B.; Lang, Y.; et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517.
- Hirst, G.K. The agglutination of red cells by allantoic fluid of chick embryos infected with influenza virus. Science 1941, 94, 22–23.
- Hirst, G.K. Adsorption of influenza hemagglutinins and virus by red blood cells. J. Exp. Med. 1942, 76, 195–209.
- McClelland, L.; Hare, R. The adsorption of influenza virus by red cells and a new in vitro method of measuring antibodies for influenza virus. Can. Public Health J. 1941, 32, 530–538.
- Salk, J.E. A Simplified procedure for titrating hemagglutinating capacity of influenza-virus and the corresponding antibody. J. Immunol. 1944, 49, 87–98.
- Salk, J.E. A plastic plate for use in tests involving virus hemagglutination and other similar reactions. Science 1948, 108, 749.
- Hierholzer, J.C.; Suggs, M.T.; Hall, E.C. Standardized viral hemagglutination and hemagglutination-inhibition tests. II. Description and statistical evaluation. Appl Microbiol 1969, 18, 824–833.
- Defang, G.N.; Martin, N.J.; Burgess, T.H.; Millar, E.V.; Pecenka, L.A.; Danko, J.R.; Arnold, J.C.; Kochel, T.J.; Luke, T.C. Comparative analysis of hemagglutination inhibition titers generated using temporally matched serum and plasma samples. PLoS ONE 2012, 7, e48229.
- Nguyen, M.; Fries, K.; Khoury, R.; Zheng, L.; Hu, B.; Hildreth, S.W.; Parkhill, R.; Warren, W. Automated imaging and analysis of the hemagglutination inhibition assay. J. Lab. Autom. 2015, 21, 287–296.
- Killian, M.L. Hemagglutination assay for influenza virus. In Animal Influenza Virus; Spackman, E., Ed.; Springer: New York, NY, USA, 2014; pp. 3–9.
- Barrett, T.; Inglis, S. Growth, purification and titration of influenza viruses. In Virology: A Practical Approach; Mahy, Ed.; Oxford IRL Press Ltd.: Oxford, UK, 1985; pp. 119–150.
- Ryu, W.-S. Chapter 4—Diagnosis and methods. In Molecular Virology of Human Pathogenic Viruses; Ryu, W.-S., Ed.; Academic Press: Boston, MA, USA, 2017; pp. 47–62.
- Pedersen, J.C. Hemagglutination-inhibition assay for influenza virus subtype identification and the detection and quantitation of serum antibodies to influenza Virus. In Animal Influenza Virus; Spackman, E., Ed.; Springer: New York, NY, USA, 2014; pp. 11–25.
- Bakkers, M.J.G.; Lang, Y.; Feitsma, L.J.; Hulswit, R.J.G.; de Poot, S.A.H.; van Vliet, A.L.W.; Margine, I.; de Groot-Mijnes, J.D.F.; van Kuppeveld, F.J.M.; Langereis, M.A.; et al. Betacoronavirus adaptation to humans involved progressive loss of hemagglutinin-esterase lectin activity. Cell Host Microbe 2017, 21, 356–366.
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.J.; Bosch, B.J.; et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690.
- Neu, U.; Bauer, J.; Stehle, T. Viruses and sialic acids: Rules of engagement. Curr. Opin. Struct. Biol. 2011, 21, 610–618.
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human Coronavirus HKU1 spike protein uses o-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015, 89, 7202–7213.
- Dai, X.; Zhang, X.; Ostrikov, K.; Abrahamyan, L. Host receptors: The key to establishing cells with broad viral tropism for vaccine production. Crit. Rev. Microbiol. 2020, 46, 147–168.
- Newton, R.; Delguste, M.; Koehler, M.; Dumitru, A.C.; Laskowski, P.R.; Müller, D.J.; Alsteens, D. Combining confocal and atomic force microscopy to quantify single-virus binding to mammalian cell surfaces. Nat. Protoc. 2017, 12, 2275–2292.
- Park, Y.-J.; Walls, A.C.; Wang, Z.; Sauer, M.M.; Li, W.; Tortorici, M.A.; Bosch, B.-J.; DiMaio, F.; Veesler, D. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 2019, 26, 1151–1157.
- Sieben, C.; Kappel, C.; Zhu, R.; Wozniak, A.; Rankl, C.; Hinterdorfer, P.; Grubmüller, H.; Herrmann, A. Influenza virus binds its host cell using multiple dynamic interactions. Proc. Natl. Acad. Sci. USA 2012, 109, 13626–13631.
- Tiralongo, J. Chapter 29—Sialic acid-specific microbial lectins. In Microbial Glycobiology; Holst, O., Brennan, P.J., Itzstein, M.V., Moran, A.P., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 585–598.
- Wielgat, P.; Rogowski, K.; Godlewska, K.; Car, H. Coronaviruses: Is Sialic acid a gate to the eye of cytokine storm? From the entry to the effects. Cells 2020, 9, 1963.
- Nelson, D.S. Immune adherence. In Advances in Immunology; Dixon, F.J., Humphrey, J.H., Eds.; Academic Press: Cambridge, MA, USA, 1963; Volume 3, pp. 131–180.
- Anderson, H.L.; Brodsky, I.E.; Mangalmurti, N.S. The evolving erythrocyte: Red blood cells as modulators of innate immunity. J. Immunol. 2018, 201, 1343–1351.
- Varki, A.; Gagneux, P. Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 2012, 1253, 16–36.
- Aoki, T. A comprehensive review of our current understanding of red blood cell (RBC) glycoproteins. Membranes 2017, 7, 56.
- Viitala, J.; Järnefelt, J. The red cell surface revisited. Trends Biochem. Sci. 1985, 10, 392–395.
- De Back, D.Z.; Kostova, E.; Klei, T.; Beuger, B.; van Zwieten, R.; Kuijpers, T.; Juffermans, N.; van den Berg, T.; Korte, D.; van Kraaij, M.; et al. RBC Adhesive Capacity Is Essential for Efficient ‘Immune Adherence Clearance’ and Provide a Generic Target to Deplete Pathogens from Septic Patients. Blood 2016, 128, 1031.
- Nelson, R.A. The Immune-Adherence Phenomenon: An Immunologically Specific Reaction Between Microorganisms and Erythrocytes Leading to Enhanced Phagocytosis. Science 1953, 118, 733–737.
- Nelson, R.A., Jr. The immune-adherence phenomenon; a hypothetical role of erythrocytes in defence against bacteria and viruses. Proc. R Soc. Med. 1956, 49, 55–58.
- de Groot, R.J.; Baker, S.C.; Baric, R.; Enjuanes, L. Family—Coronaviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2012; pp. 806–828.
- Sriwilaijaroen, N.; Suzuki, Y. Sialoglycovirology of lectins: Sialyl Glycan binding of enveloped and non-enveloped viruses. In Lectin Purification and Analysis: Methods and Protocols; Hirabayashi, J., Ed.; Springer: New York, NY, USA, 2020; pp. 483–545.
- Lang, Y.; Li, W.; Li, Z.; Koerhuis, D.; van den Burg, A.C.S.; Rozemuller, E.; Bosch, B.-J.; van Kuppeveld, F.J.M.; Boons, G.-J.; Huizinga, E.G.; et al. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc. Natl. Acad. Sci. USA 2020, 117, 25759–25770.
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033.
- Awasthi, M.; Gulati, S.; Sarkar, D.; Tiwari, S.; Kateriya, S.; Ranjan, P.; Verma, S.K. The Sialoside-Binding Pocket of SARS-CoV-2 Spike Glycoprotein Structurally Resembles MERS-CoV. Viruses 2020, 12, 909.
- Chen, W.; Hui, Z.; Ren, X.; Luo, Y.; Shu, J.; Yu, H.; Li, Z. The N-glycosylation sites and Glycan-binding ability of S-protein in SARS-CoV-2 Coronavirus. bioRxiv 2020, 12.01.406025.
- Guo, W.; Lakshminarayanan, H.; Rodriguez-Palacios, A.; Salata, R.A.; Xu, K.; Draz, M.S. Glycan Nanostructures of Human Coronaviruses. Int. J. Nanomed. 2021, 16, 4813–4830.
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988.
- Gao, C.; Zeng, J.; Jia, N.; Stavenhagen, K.; Matsumoto, Y.; Zhang, H.; Li, J.; Hume, A.J.; Mühlberger, E.; van Die, I.; et al. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv 2020, 07.29.227462.
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236.
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423.
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820.
- Kumar, S.; Nyodu, R.; Maurya, V.K.; Saxena, S.K. Morphology, Genome Organization, Replication, and Pathogenesis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). In Coronavirus Disease 2019 (COVID-19): Epidemiology, Pathogenesis, Diagnosis, and Therapeutics; Saxena, S.K., Ed.; Springer: Singapore, 2020; pp. 23–31.
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216.
- Bakkers, M.J.G.; Zeng, Q.; Feitsma, L.J.; Hulswit, R.J.G.; Li, Z.; Westerbeke, A.; van Kuppeveld, F.J.M.; Boons, G.-J.; Langereis, M.A.; Huizinga, E.G.; et al. Coronavirus receptor switch explained from the stereochemistry of protein–carbohydrate interactions and a single mutation. Proc. Natl. Acad. Sci. USA 2016, 113, E3111–E3119.
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A.; et al. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020, 6, 2046–2052.
- Collins, B.E.; Paulson, J.C. Cell surface biology mediated by low affinity multivalent protein–glycan interactions. Curr. Opin. Chem. Biol. 2004, 8, 617–625.
- Cohen, M.; Varki, A. Chapter Three—Modulation of Glycan Recognition by Clustered Saccharide Patches. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 308, pp. 75–125.
- Xiong, X.; Coombs, P.J.; Martin, S.R.; Liu, J.; Xiao, H.; McCauley, J.W.; Locher, K.; Walker, P.A.; Collins, P.J.; Kawaoka, Y.; et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 2013, 497, 392–396.
- Xu, H.; Shaw, D.E. A simple model of multivalent adhesion and its application to influenza infection. Biophys. J. 2016, 110, 218–233.
- Sauter, N.K.; Hanson, J.E.; Glick, G.D.; Brown, J.H.; Crowther, R.L.; Park, S.J.; Skehel, J.J.; Wiley, D.C. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: Analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography. Biochemistry 1992, 31, 9609–9621.
- Mitnaul, L.J.; Matrosovich, M.N.; Castrucci, M.R.; Tuzikov, A.B.; Bovin, N.V.; Kobasa, D.; Kawaoka, Y. Balanced Hemagglutinin and Neuraminidase Activities Are Critical for Efficient Replication of Influenza A Virus. J. Virol. 2000, 74, 6015–6020.
- Pourrajab, F.; Zare-Khormizi, M.R.; Sheikhha, M.H. Molecular basis for pathogenicity of human coronaviruses. Infect. Drug Resist. 2020, 13, 2385–2405.
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020, 75, 2829–2845.
- Silva-Filho, J.C.; Melo, C.G.F.d.; Oliveira, J.L.d. The influence of ABO blood groups on COVID-19 susceptibility and severity: A molecular hypothesis based on carbohydrate-carbohydrate interactions. Med. Hypotheses 2020, 144, 110155.
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal. Transduct. Target. Ther. 2020, 5, 283.
- Bian, H.; Zheng, Z.-H.; Wei, D.; Wen, A.; Zhang, Z.; Lian, J.-Q.; Kang, W.-Z.; Hao, C.-Q.; Wang, J.; Xie, R.-H.; et al. Safety and efficacy of meplazumab in healthy volunteers and COVID-19 patients: A randomized phase 1 and an exploratory phase 2 trial. Signal. Transduct. Target. Ther. 2021, 6, 194.
- Shelokov, A.; Vogel, J.E.; Chi, L. Hemadsorption (Adsorption-Hemagglutination) Test for Viral Agents in Tissue Culture with Special Reference to Influenza. Proc. Soc. Exp. Biol. Med. 1958, 97, 802–809.
- Gagneten, S.; Gout, O.; Dubois-Dalcq, M.; Rottier, P.; Rossen, J.; Holmes, K.V. Interaction of mouse hepatitis virus (MHV) spike glycoprotein with receptor glycoprotein MHVR is required for infection with an MHV strain that expresses the hemagglutinin-esterase glycoprotein. J. Virol. 1995, 69, 889–895.
- Adams, J.H.; Sim, B.K.; Dolan, S.A.; Fang, X.; Kaslow, D.C.; Miller, L.H. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA 1992, 89, 7085–7089.
- Persson, K.E.; McCallum, F.J.; Reiling, L.; Lister, N.A.; Stubbs, J.; Cowman, A.F.; Marsh, K.; Beeson, J.G. Variation in use of erythrocyte invasion pathways by Plasmodium falciparum mediates evasion of human inhibitory antibodies. J. Clin. Invest. 2008, 118, 342–351.
- Crosnier, C.; Bustamante, L.Y.; Bartholdson, S.J.; Bei, A.K.; Theron, M.; Uchikawa, M.; Mboup, S.; Ndir, O.; Kwiatkowski, D.P.; Duraisingh, M.T.; et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 2011, 480, 534–537.
- Zenonos, Z.A.; Dummler, S.K.; Müller-Sienerth, N.; Chen, J.; Preiser, P.R.; Rayner, J.C.; Wright, G.J. Basigin is a druggable target for host-oriented antimalarial interventions. J. Exp. Med. 2015, 212, 1145–1151.
- Berzuini, A.; Bianco, C.; Migliorini, A.C.; Maggioni, M.; Valenti, L.; Prati, D. Red blood cell morphology in patients with COVID-19-related anaemia. Blood Transfus. 2021, 19, 34–36.
- Lakhdari, N.; Tabet, B.; Boudraham, L.; Laoussati, M.; Aissanou, S.; Beddou, L.; Bensalem, S.; Bellik, Y.; Bournine, L.; Fatmi, S.; et al. Red blood cells injuries and hypersegmented neutrophils in COVID-19 peripheral blood film. medRxiv 2020, 07.24.20160101.
- Melkumyants, A.; Buryachkovskaya, L.; Lomakin, N.; Antonova, O.; Serebruany, V. Mild COVID-19 and impaired blood cell–endothelial crosstalk: Considering long-term use of antithrombotics? Thromb. Haemost. 2022, 122, 123–130.
- Diez-Silva, M.; Dao, M.; Han, J.; Lim, C.-T.; Suresh, S. Shape and Biomechanical Characteristics of Human Red Blood Cells in Health and Disease. MRS Bull. 2010, 35, 382–388.
- Guest, M.M.; Bond, T.P.; Cooper, R.G.; Derrick, J.R. Red Blood Cells: Change in Shape in Capillaries. Science 1963, 142, 1319–1321.
- Shahid, M.; Nunhuck, A. Physiology; Elsevier Health Sciences: Philadelphia, PA, USA, 2008; p. 273.
- Kuchel, P.W.; Shishmarev, D. Accelerating metabolism and transmembrane cation flux by distorting red blood cells. Sci. Adv. 2017, 3, EAAO1016.
- Ocak, I.; Kara, A.; Ince, C. Monitoring microcirculation. Best Pract. Res. Clin. Anaesthesiol. 2016, 30, 407–418.
- Chasis, J.A.; Mohandas, N. Red blood cell glycophorins. Blood 1992, 80, 1869–1879.
- Barshtein, G.; Wajnblum, D.; Yedgar, S. Kinetics of linear rouleaux formation studied by visual monitoring of red cell dynamic organization. Biophys. J. 2000, 78, 2470–2474.
- Fung, Y.-C. The flow properties of blood. In Biomechanics: Mechanical Properties of Living Tissues; Fung, Y.-C., Ed.; Springer New York: New York, NY, USA, 1993; pp. 66–108.
- Maeda, N.; Seike, M.; Kon, K.; Shiga, T. Erythrocyte Aggregation as a Determinant of Blood Flow: Effect of pH, Temperature and Osmotic Pressure. In Oxygen Transport to Tissue X; Mochizuki, M., Honig, C.R., Koyama, T., Goldstick, T.K., Bruley, D.F., Eds.; Springer: New York, NY, USA, 1988; pp. 563–570.
- Sakariassen, K.S.; Orning, L.; Turitto, V.T. The impact of blood shear rate on arterial thrombus formation. Future Sci. OA 2015, 1, FSO30.
- Ahmetaj-Shala, B.; Vaja, R.; Atanur, S.; George, P.; Kirkby, N.; Mitchell, J. Systemic analysis of putative SARS-CoV-2 entry and processing genes in cardiovascular tissues identifies a positive correlation of BSG with age in endothelial cells. bioRxiv 2020, 06.23.165324.
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128.
- Siddiqi, H.; Libby, P.; Ridker, P. COVID-19 and vascular disease. EBioMedicine 2020, 58, 102966.
- Lee, M.-H.; Perl, D.P.; Nair, G.; Li, W.; Maric, D.; Murray, H.; Dodd, S.J.; Koretsky, A.P.; Watts, J.A.; Cheung, V.; et al. Microvascular Injury in the Brains of Patients with Covid-19. N. Engl. J. Med. 2020, 384, 481–483.
- Colunga Biancatelli, R.M.L.; Solopov, P.A.; Sharlow, E.R.; Lazo, J.S.; Marik, P.E.; Catravas, J.D. The SARS-CoV-2 spike protein subunit S1 induces COVID-19-like acute lung injury in Κ18-hACE2 transgenic mice and barrier dysfunction in human endothelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2021, 321, L477–L484.
- Nuovo, G.J.; Magro, C.; Shaffer, T.; Awad, H.; Suster, D.; Mikhail, S.; He, B.; Michaille, J.-J.; Liechty, B.; Tili, E. Endothelial cell damage is the central part of COVID-19 and a mouse model induced by injection of the S1 subunit of the spike protein. Ann. Diagn. Pathol. 2021, 51, 151682.
- Perico, L.; Morigi, M.; Galbusera, M.; Pezzotta, A.; Gastoldi, S.; Imberti, B.; Ruggenenti, P.; Benigni, A.; Remuzzi, G. SARS-CoV-2 Spike Protein 1 Activates Microvascular Endothelial Cells and Complement System Leading to Thrombus Formation. Available online: https://ssrn.com/abstract=3864027 (accessed on 23 November 2021).
- Magro, C.; Mulvey, J.J.; Laurence, J.; Sanders, S.; Crowson, N.; Grossman, M.; Harp, J.; Nuovo, G. The differing pathophysiologies that underlie COVID-19 associated perniosis and thrombotic retiform purpura: A case series. Br. J. Derm. 2020, 184, 141–150.
- Magro, C.M.; Mulvey, J.J.; Laurence, J.; Seshan, S.; Crowson, A.N.; Dannenberg, A.J.; Salvatore, S.; Harp, J.; Nuovo, G.J. Docked severe acute respiratory syndrome coronavirus 2 proteins within the cutaneous and subcutaneous microvasculature and their role in the pathogenesis of severe coronavirus disease 2019. Hum. Pathol. 2020, 106, 106–116.
- Ko, C.J.; Harigopal, M.; Gehlhausen, J.R.; Bosenberg, M.; McNiff, J.M.; Damsky, W. Discordant anti-SARS-CoV-2 spike protein and RNA staining in cutaneous perniotic lesions suggests endothelial deposition of cleaved spike protein. J. Cutan Pathol 2021, 48, 47–52.
- Bernard, I.; Limonta, D.; Mahal, L.K.; Hobman, T.C. Endothelium Infection and Dysregulation by SARS-CoV-2: Evidence and Caveats in COVID-19. Viruses 2021, 13, 29.
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734.
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330.
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261.
- Weiss, A.; Jellingsø, M.; Sommer, M.O.A. Spatial and temporal dynamics of SARS-CoV-2 in COVID-19 patients: A systematic review and meta-analysis. EBioMedicine 2020, 58, 102916.
- Sikora, M.; von Bülow, S.; Blanc, F.E.C.; Gecht, M.; Covino, R.; Hummer, G. Computational epitope map of SARS-CoV-2 spike protein, Supplementary Information, Movie S1. PLoS Comput. Biol. 2021, 17, e1008790.
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein; supplementary Information. ACS Cent. Sci. 2020, 6, 1722–1734.
- Beniac, D.R.; Andonov, A.; Grudeski, E.; Booth, T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006, 13, 751–752.
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502.
- Kiss, B.; Kis, Z.; Pályi, B.; Kellermayer, M.S.Z. Topography, Spike Dynamics, and Nanomechanics of Individual Native SARS-CoV-2 Virions. Nano Lett. 2021, 21, 2675–2680.
- Laue, M.; Kauter, A.; Hoffmann, T.; Möller, L.; Michel, J.; Nitsche, A. Morphometry of SARS-CoV and SARS-CoV-2 particles in ultrathin plastic sections of infected Vero cell cultures. Sci. Rep. 2021, 11, 3515.
- Huang, Y.; Yang, C.; Xu, X.-f.; Xu, W.; Liu, S.-w. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149.
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263.
- Chabert, A.; Hamzeh-Cognasse, H.; Pozzetto, B.; Cognasse, F.; Schattner, M.; Gomez, R.M.; Garraud, O. Human platelets and their capacity of binding viruses: Meaning and challenges? BMC Immunol. 2015, 16, 26.
- Pryzdial, E.L.G.; Lin, B.H.; Sutherland, M.R. Virus–Platelet Associations. In Platelets in Thrombotic and Non-Thrombotic Disorders: Pathophysiology, Pharmacology and Therapeutics: An Update; Gresele, P., Kleiman, N.S., Lopez, J.A., Page, C.P., Eds.; Springer International Publishing: New York, NY, USA, 2017; pp. 1085–1102.
- Stocker, T.J.; Ishikawa-Ankerhold, H.; Massberg, S.; Schulz, C. Small but mighty: Platelets as central effectors of host defense. Thromb. Haemost. 2017, 117, 651–661.
- Hyvärinen, S.; Meri, S.; Jokiranta, T.S. Disturbed sialic acid recognition on endothelial cells and platelets in complement attack causes atypical hemolytic uremic syndrome. Blood 2016, 127, 2701–2710.
- Nissilä, E.; Hakala, P.; Leskinen, K.; Roig, A.; Syed, S.; Van Kessel, K.P.M.; Metso, J.; De Haas, C.J.C.; Saavalainen, P.; Meri, S.; et al. Complement Factor H and Apolipoprotein E Participate in Regulation of Inflammation in THP-1 Macrophages. Front. Immunol. 2018, 9, 2701.
- Soerensen, A.L.; Wandall, H.H.; Patel, S.; Richardson, J.; Italiano, J.; Clausen, H.; Stossel, T.P.; Hartwig, J.H.; Hoffmeister, K.M. Platelets Lacking Sialic Acid Clear Rapidly from the Circulation Due to Ingestion by Asialoglycoprotein Receptor-Expressing Liver Macrophages and Hepatocytes. Blood 2006, 108, 1521.
- Assinger, A. Platelets and Infection—An Emerging Role of Platelets in Viral Infection. Front. Immunol. 2014, 5, 649.
- Kasinrerk, W.; Tokrasinwit, N.; Phunpae, P. CD147 monoclonal antibodies induce homotypic cell aggregation of monocytic cell line U937 via LFA-1/ICAM-1 pathway. Immunology 1999, 96, 184–192.
- Pennings, G.J.; Kritharides, L. CD147 in cardiovascular disease and thrombosis. Semin. Thromb. Hemost. 2014, 40, 747–755.
- Loh, D. The potential of melatonin in the prevention and attenuation of oxidative hemolysis and myocardial injury from cd147 SARS-CoV-2 spike protein receptor binding. Melatonin Res. 2020, 3, 380–416.
- Pushkarsky, T.; Zybarth, G.; Dubrovsky, L.; Yurchenko, V.; Tang, H.; Guo, H.; Toole, B.; Sherry, B.; Bukrinsky, M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 2001, 98, 6360–6365.
- Rowe, J.A.; Claessens, A.; Corrigan, R.A.; Arman, M. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: Molecular mechanisms and therapeutic implications. Expert Rev. Mol. Med. 2009, 11, e16.
- Adams, Y.; Kuhnrae, P.; Higgins, M.K.; Ghumra, A.; Rowe, J.A. Rosetting Plasmodium falciparum-infected erythrocytes bind to human brain microvascular endothelial cells in vitro, demonstrating a dual adhesion phenotype mediated by distinct P. falciparum erythrocyte membrane protein 1 domains. Infect. Immun. 2014, 82, 949–959.
- Arman, M.; Rowe, J.A. Experimental conditions affect the outcome of Plasmodium falciparum platelet-mediated clumping assays. Malar. J. 2008, 7, 243.
- Deroost, K.; Pham, T.T.; Opdenakker, G.; Van den Steen, P.E. The immunological balance between host and parasite in malaria. FEMS Microbiol. Rev. 2016, 40, 208–257.
- Choi, Y.K.; Cao, Y.; Frank, M.; Woo, H.; Park, S.-J.; Yeom, M.S.; Croll, T.I.; Seok, C.; Im, W. Structure, Dynamics, Receptor Binding, and Antibody Binding of the Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein in a Viral Membrane. J. Chem. Theory Comput. 2021, 17, 2479–2487.
- Lardone, R.D.; Garay, Y.C.; Parodi, P.; de la Fuente, S.; Angeloni, G.; Bravo, E.O.; Schmider, A.K.; Irazoqui, F.J. How glycobiology can help us treat and beat the COVID-19 pandemic. J. Biol. Chem. 2021, 296, 100375.
- Bharara, R.; Singh, S.; Pattnaik, P.; Chitnis, C.E.; Sharma, A. Structural analogs of sialic acid interfere with the binding of erythrocyte binding antigen-175 to glycophorin A, an interaction crucial for erythrocyte invasion by Plasmodium falciparum. Mol. Biochem. Parasitol. 2004, 138, 123–129.
- Jaskiewicz, E.; Jodłowska, M.; Kaczmarek, R.; Zerka, A. Erythrocyte glycophorins as receptors for Plasmodium merozoites. Parasites Vectors 2019, 12, 317.
- Halbhuber, K.J.; Gliesing, M.; Stibenz, D.; Makovitzky, J. Topo-optical investigations of the human erythrocyte glycocalyx-age related changes. Histochemistry 1984, 81, 187–193.
- van Oss, C.J.; Absolom, D.R. Zeta potentials, van der Waals forces and hemagglutination. Vox Sang. 1983, 44, 183–190.
- Pretini, V.; Koenen, M.H.; Kaestner, L.; Fens, M.H.A.M.; Schiffelers, R.M.; Bartels, M.; Van Wijk, R. Red Blood Cells: Chasing Interactions. Front. Physiol. 2019, 10.
- Fernandes, H.P.; Cesar, C.L.; Barjas-Castro Mde, L. Electrical properties of the red blood cell membrane and immunohematological investigation. Rev. Bras. Hematol Hemoter 2011, 33, 297–301.





