ARoulthougheaux (stacked clumps) of red blood cells (RBCs) observed in the blood of COVID-19 typically gains infectious penetration in the respiratory epithelium, vascular damage is frequently observed in lungs and other organ systemspatients in three studies call attention to the properties of several enveloped virus strains dating back to seminal findings of the 1940s. For COVID-19, key such properties are: (1) SARS-CoV-2 binds to RBCs in vitro and also in the blood of COVID-19 patients, with morbidities such as intravascular clotting, microvascular occlusion and peripheral ischemia; (2) although ACE2 is its target for viral fusion and replication, SARS-CoV-2 initially attaches to sialic acid (SA) terminal moieties on host cell membranes via glycans on its spike protein; (3) certain enveloped viruses express hemagglutinin esterase (HE), an enzyme that releases these glycan-mediated bindings to host cells, which is expressed among betacoronaviruses in the common cold strains but not the virulent strains, SARS-CoV, SARS-CoV-2 and MERS. The arrangement and chemical composition of the glycans at the 22 N-glycosylation sites of SARS-CoV-2 spike protein and those at the sialoglycoprotein coating of RBCs allow exploration of specifics as to how virally induced RBC clumping may form. The in vitro and clinical testing of these possibilities can be sharpened by the incorporation of an existing anti-COVID-19 therapeutic that has been found in silico to competitively bind to multiple glycans on SARS-CoV-2 spike protein.
1. Introduction
Although COVID-19 typically gains infectious penetration in the respiratory epithelium
[1][2][3][1,2,3], vascular damage is frequently observed in lungs and other organ systems of COVID-19 patients, with morbidities such as intravascular clotting, microvascular occlusion and peripheral ischemia
[4][5][6][7][8][9][10][11][12][13][14][15][16][17][4,5,6,7,8,9,10,11,12,13,14,15,16,17]. Histological studies from COVID-19 patients have found extensively damaged endothelium of pulmonary capillaries adjoining relatively intact alveoli
[18][19][18,19], corresponding to hypoxemia with normal breathing mechanics observed in patients with this viral disease
[10][16][17][18][20][21][10,16,17,18,20,21]. One clinical reviewer characterized COVID-19 as “a systemic disease that primarily injures the vascular endothelium”
[21].
A framework for studying the vascular occlusive morbidities characteristic of COVID-19 is provided by viral properties dating back to classic experiments of the 1940s, as reviewed
[22]. Viruses fuse and then replicate via host cell receptors specific to the viral strain, which is ACE2 for SARS-CoV-2
[23][24][23,24]. However, many enveloped viral strains, including coronaviruses, initially attach to host cell membranes via glycoconjugate molecules, including those tipped with sialic acid (SA)
[25][26][27][28][29][30][25,26,27,28,29,30]. SA is densely distributed on red blood cells (RBCs) as terminal residues of its surface sialoglycoprotein, glycophorin A (GPA), and of its CD147 transmembrane receptors
[31][32][33][34][31,32,33,34]. Through viral bindings to SA surface moieties to be considered in detail below, SARS-CoV-2 agglutinates RBCs, as established in vitro
[35], with such bindings also demonstrated clinically
[36]. Hemagglutination also occurs for several other viral strains including other coronaviruses
[35][37][38][39][40][41][42][43][44][45][46][47][48][35,37,38,39,40,41,42,43,44,45,46,47,48], as demonstrated in the classic viral hemagglutination assay. In that assay, developed in the 1940s
[38][49][50][51][38,49,50,51] and refined later by Jonas Salk
[52][53][54][55][56][52,53,54,55,56], virus particles are mixed with RBCs to form a hemagglutinated sheet
[40][57][58][59][40,57,58,59]. However, for viruses that express enzymes that cleave SA, that sheet subsequently collapses as these enzymes disintegrate SA-binding sites on the RBCs
[38][58][60][61][38,58,60,61].
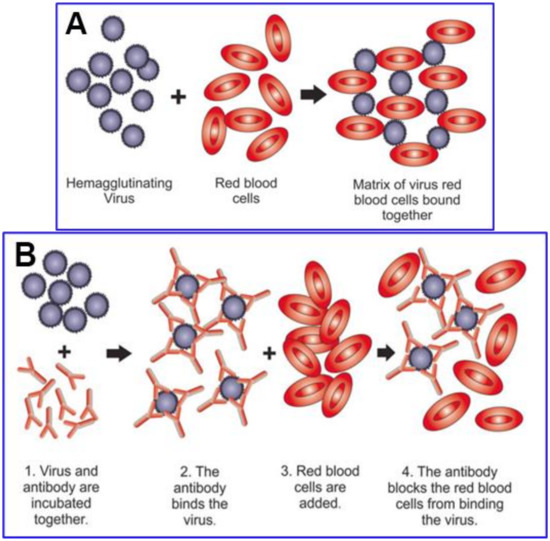
Figure 1 shows a schematic of virally mediated hemagglutination and of its inhibition through an agent that competitively binds to attachment sites on the virus, in this case an antiviral antibody.
Figure 1. Schematic of hemagglutination (
A) and hemagglutination inhibition (
B), both of which occur either in vitro in their respective assays or in vivo. Reproduced with permission from Springer Nature ((
A) Killian, 2014
[57]; (
B) Pedersen, 2014
[60]). (
B) depicts blockage of hemagglutination by an antibody to the virus.
1.1. Binding of Viruses to SA and Host Decoy Defense
For viruses that bind to SA, including SARS-CoV-2 as noted above, such glycan bindings play a key role in viral infectivity, as SA typically functions as an initial point of attachment to host cells
[25][30][44][48][62][63][64][65][66][67][68][69][70][25,30,44,48,62,63,64,65,66,67,68,69,70]. The human host, reciprocally, protects against attachment of virions to host cell infectious targets by presenting SA on a set of decoys, including RBCs as well as platelets and leukocytes
[22][31][71][72][73][22,31,71,72,73]. In particular, SA moieties are densely distributed on the RBC surface (35 million per cell
[33]), mainly as terminal residues of both the CD147 receptor
[22] and GPA
[31][74][31,74], with GPA serving no known physiological role other than as a decoy for pathogens
[31][72][73][75][31,72,73,75]. RBCs perform an active pathogen clearance role, attaching to microorganisms and then delivering them to leukocytes or conveying them to macrophages in the liver or spleen for phagocytosis
[31][71][73][76][31,71,73,76]. With bacteria, this clearance by RBCs requires both antibody and complement
[77][78][77,78]. Viruses, however, are snagged directly by RBCs, and viral–RBC binding is inhibited by antiviral antibodies
[22]. RBCs are well suited for this immune defense role, having no nucleus or other infrastructure to perform viral replication and being expendable, numbering 20–30 trillion per human host
[72].
1.2. Cleavage of Viral–SA Binding via Hemagglutinin Esterase (HE), an Enzyme Expressed by the Common Cold Betacoronavirus Strains but Not by SARS-CoV, SARS-CoV-2 and MERS
Hemagglutinin esterase (HE) is an SA-cleaving enzyme deployed by certain viral strains, including some betacoronaviruses. HE increases the infectivity of these viral strains by expediting the release of replicated virions from sialoside-binding sites on host cells and by restricting viral snagging on such binding sites of non-infectious targets, including mucins, blood cells and plasma proteins
[22]. Viral HE can both bond and cleave host cell sialoside-binding sites
[79][80][79,80]; however, for betacoronaviruses, even for those expressing HE, attachments to host cell sialoside-binding sites are effected mainly through glycans at 22 N-glycosylation sites on viral spike protein S1, in particular, eight of those clustered on the N-terminal domain (NTD)
[44][81][82][83][84][85][86][87][44,81,82,83,84,85,86,87]. Genomic analysis of the five strains of human betacoronaviruses reveals expression of the HE enzyme in those that cause the common cold, OC43 and HKU1, but not in the three deadly strains, SARS-CoV, SARS-CoV-2 and MERS
[88][89][90][91][92][88,89,90,91,92].
1.3. Multivalent Binding
To adjust for a missing release capability in the three betacoronavirus strains lacking HE, SA-binding affinity is diminished, while multiplicity of binding provides robust variations in attachment strength
[44][48][62][63][93][94][44,48,62,63,93,94]. These viral bonds to SA and other glycans have weak individual affinities but greatly increased collective strength
[25][30][44][48][63][66][93][94][95][96][25,30,44,48,63,66,93,94,95,96]. The dissociation constant Kd for a multivalent attachment of viral spike protein to SA increases exponentially (in absolute value) as a function of the number of bonds
[97][98][97,98]. For example, the Kd of an individual bond of viral spike protein to a host sialoside-binding site is in the low millimolar range
[63][93][99][100][63,93,99,100], but the Kd value would be in the nanomolar range for a triple bond
[97]. A single sialoside bond for a virus particle initially attaching to a cell would provide only a tentative foothold
[25][30][44][63][101][102][25,30,44,63,101,102], allowing migration to an ACE2 receptor for fusion and replication.
2. Properties of SARS-CoV-2 Related to Its Array of Spike Glycoprotein Glycans
2.1. SARS-CoV-2 Binds to SA and CD147
Because bindings from viral spike protein to SA are generally weak when univalent, several coronavirus strains require a nanoarray experimental detection methodology to register viral attachment to hosts
[22][48][61][62][22,48,61,62]. Using such a methodology, a nanoparticle array bearing SA derivatives, the binding of both SARS-CoV-2 spike protein and pseudovirus was demonstrated
[94]. This
enst
rudy’s detection system is adaptable to mass COVID-19 screening, with a 5 nM concentration threshold for detection of viral spike protein. Another attachment point on host cells for the SARS-CoV-2 virus is the CD147 transmembrane receptor, which contains SA at its terminal domains
[34][103][34,103]. Binding of SARS-CoV-2 spike protein to CD147 was shown by enzyme-linked immunosorbent assay (ELISA), surface plasmon resonance (SPR) and co-immunoprecipitation (Co-IP) assays, and colocalization of CD147 and viral spike protein was revealed on infected Vero E6 cells
[104]. Meplazumab, a humanized anti-CD147 antibody, was found to inhibit viral proliferation in vitro and to provide significant clinical benefits in a small study of its use for COVID-19 treatment
[105].
2.2. SARS-CoV-2 Attaches to RBCs, Other Blood Cells and Endothelial Cells
Several of the glycans at the 22 N-glycosylation sites and both O-linked glycans on SARS-CoV-2 spike protein (as depicted in Figure 3) are capped with SA monosaccharides of the same type that are densely distributed on human RBCs at the tips of GPA molecules, as will be considered in some detail below. These matching glycans enable SARS-CoV-2 to hemagglutinate when mixed with human RBCs, as indeed demonstrated using the hemadsorption assay
[35] (similar to the hemagglutination assay
[106][107][106,107]). Hemagglutination occurs more generally in eight families of viruses, including other coronaviruses
[35][37][38][39][40][41][42][43][44][45][46][47][48][35,37,38,39,40,41,42,43,44,45,46,47,48]. Attachment of SARS-CoV-2 spike protein to RBCs was demonstrated directly through immunofluorescence analysis of RBCs from the blood of nine hospitalized COVID-19 patients
[36]. The mean percentage of RBCs having SARS-CoV-2 spike protein punctae was 41% at day 0 of hospital admission, with values ranging from 0% for one patient and 18% for two patients to 79% for another patient. This mean percentage increased to 44% at day 7 after hospital admission.
Like RBCs, several other cell types express surface SA glycoconjugates and can thus also attach to SARS-CoV-2 virus particles. SA and SA-tipped CD147 are expressed on endothelial cells of blood vessel linings (luminal surface), platelets, lymphocytes, macrophages, and other types of white blood cells
[22]. The potential for pathogen attachments to SA and CD147 that impede vascular blood flow is indicated in another disease, severe malaria, in which the malaria parasite attaches to SA-binding sites on an RBC
[73][108][109][73,108,109] and penetrates the RBC through the latter’s CD147 receptors
[110][111][110,111]. Clumps develop between infected and uninfected RBCs, often including platelets, which, along with endothelial cytoadhesion by infected RBCs, cause vascular occlusion, the key morbidity of severe malaria
[22].
The interlaced attachments of RBCs with SARS-CoV-2 virions as observed in vitro in the hemadsorption assay
[35] may well define the mechanism by which clumps (rouleaux) of RBCs form in the blood of COVID-19 patients
[112][113][114][112,113,114], as shown in
Figure 2. These clumps would present vascular obstructive potential, given that an RBC of average disk diameter 8 µm
[115][116][115,116] traverses through an alveolar capillary of smaller average cross-sectional diameter
[117], achieved only by a distortion of the RBC’s shape to press against the capillary wall
[116][118][116,118]. Such RBC clumps could be a prime cause of the microvascular occlusion which, as noted above, is characteristic of COVID-19. These clumps could contribute as well to microvascular occlusion in larger capillaries, of cross-sectional diameter up to 20 µm
[115][119][115,119], elsewhere in the body. Vascular occlusion would be promoted even though such stacked RBC clumps (rouleaux) typically dynamically aggregate and disaggregate
[120][121][122][120,121,122], as do RBC aggregates that can form in the absence of pathogens, promoted by macromolecules in plasma under conditions of low blood shear rates
[123][124][123,124]. Formation of RBC rouleaux would increase blood viscosity
[121][122][121,122], impeding blood flow, especially in the small-diameter pulmonary capillaries, which would cascade as reduction in both flow velocity and associated shear forces would in turn tend to favor aggregation vs. disaggregation and further occlude flow
[121].
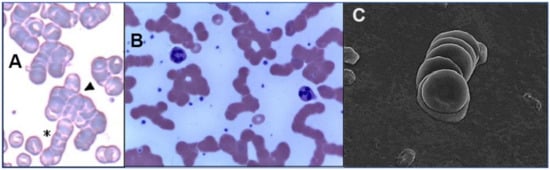
Figure 2. Images of RBC rouleaux (clumps) from the blood of COVID-19 patients, obtained using light ((
A)
[112], (
B)
[113]) and electron microscopy ((
C)
[114]). The first study (
A) found huge rouleaux formation by RBCs in 85% of COVID-19 patients studied
[112]; the second (
B) found these in 33% of patients
[113]; and the third (
C) found these prevalent in its series of 31 patients, all with mild COVID-19
[114]. Reproduced with permission from (
A) SIMTIPRO Srl; (
B) CC-BY 4.0; (
C) Georg Thieme Verlag KG.
The abundant distribution of SA-tipped CD147 on endothelial cells of blood vessel linings, with 28,000 CD147 receptors (vs. 175 ACE2 receptors) per endothelial cell
[125], may be key to the attachments of SARS-CoV-2 to endothelium and the ensuing damage that has been widely observed in COVID-19 patients
[15][18][19][126][127][128][15,18,19,126,127,128]. Damage to endothelium caused by SARS-CoV-2 spike protein in the absence of whole virus was demonstrated both in vitro and in vivo in three studies
[129][130][131][129,130,131], and presence of isolated SARS-CoV-2 spike protein on endothelial cells was also observed clinically
[130][132][133][134][135][130,132,133,134,135]. Additionally, one study of 31 hospitalized patients with mild to moderate COVID-19 found that serum levels of circulating endothelial cells (CECs), as determined by different measures, were up to 100-fold the levels for matched controls, and that these CECs from the COVID-19 patients typically each had several holes in their membranes approximately the size of SARS-CoV-2 viral capsid (the viral envelope)
[114].
2.3. The Glycan Distribution and Composition at the 22 N-Glycosylation Sites of SARS-CoV-2 Spike Protein
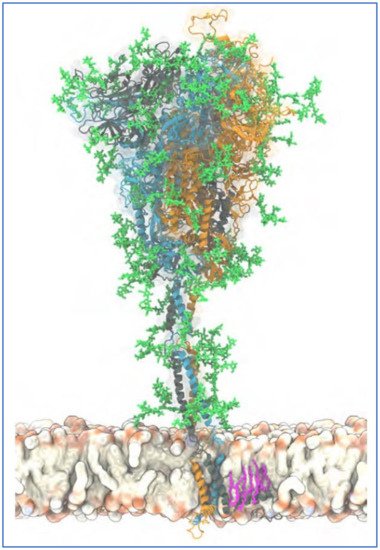
To explore key characteristics of glycan-mediated SARS-CoV-2 spike protein binding to host cells, it is helpful to consider the specific distribution and composition of the glycans at its glycosylation sites. SARS-CoV-2 spike glycoprotein is a trimer with a central helical stalk embedded in the viral envelope at its C-terminal end. The stalk consists of three joined S2 subunits each capped with an S1 subunit head spreading out in a mushroom-like shape
[136][137][138][139][136,137,138,139]. An atomistic model of a full-length trimeric SARS-CoV-2 spike protein with its attached glycans, its C-terminal (stalk) end embedded in the viral envelope, is shown in
Figure 3.
Figure 3. Atomistic model of the full-length trimeric S protein of SARS-CoV-2 shown in cartoon representation. Reproduced from Sikora et al., 2021
[140] (CC-BY 4.0). The three monomeric chains are differentiated by color. Palmitoylated cysteine residues are shown in pink licorice (only one chain shown for clarity), anchored into the viral envelope. Glycans are shown in green licorice representation. A 20 s movie showing a 600 ns atomistic molecular dynamics simulation trajectory of four S proteins embedded in a viral membrane is also provided at this source
[140].
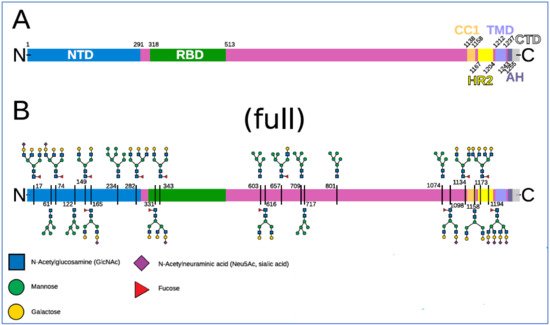
A map of the 22 N-linked glycans on each of a spike’s three monomers is shown in
Figure 4. In addition, two O-linked glycosylation sites, S325 and T323, were identified for each spike monomer, both on S1 RBD
[86], and both containing SA terminal monosaccharides
[141]. Each SARS-CoV-2 virion has a diameter, excluding spikes, of approximately 100 nm, with the number of spikes estimated at up to 65 per virion, these spikes having a length of approximately 20 nm
[142][143][144][145][142,143,144,145].
Figure 4. Spike domains and glycosylation. Reproduced from Sikora et al., 2021
[140] (CC-BY 4.0). (
A) Domains of S (SARS-CoV-2 spike protein). (
B) Glycosylation pattern of S. Sequons are indicated with the respective glycans in a schematic representation for a fully glycosylated system (“full”). A key to the monosaccharides represented is shown at the bottom.
The NTD on SARS-CoV-2 spike protein S1, at its N-terminal end, is a focal region for the spike’s glycans—eight of the spike’s 22 N-glycosylation sites are located there
[84][85][86][84,85,86]. The NTD is accordingly the typical point of initial viral attachments to glycoconjugate binding sites on host cells
[44][81][82][83][84][85][86][87][44,81,82,83,84,85,86,87]. After initial attachment, viral fusion to a host cell begins with linkage of the spike’s receptor-binding domain (RBD), situated just below NTD on spike S1, to an ACE2 receptor on the host cell membrane. The S2 stalk then becomes engaged and viral replication proceeds
[47][138][146][47,138,146]. The RBD, one on each of a spike’s three monomers, constantly switches between open (“up”) and closed (“down”) configurations, the former enabling both ACE2 binding and immune surveillance, the latter blocking both of those functions
[136][147][136,147].
ReseaOur
chers' focus now shifts to specifics of virally-mediated clumping of RBCs, notwithstanding the importance of inflammatory processes and endothelial damage in triggering and exacerbating the morbidities of COVID-19, especially in its critical phase. Additionally, platelets, the second most copious blood component
[147], serve a pathogen clearance role like that of RBCs
[148][149][150][148,149,150], and like RBCs have abundant CD147 and SA surface molecules
[151][152][153][154][155][156][157][158][151,152,153,154,155,156,157,158]. Platelets can adhere to viruses, RBCs and endothelial cells, especially under inflammatory conditions
[16][22][149][16,22,149], and are often enmeshed in clumps that develop in severe malaria between infected and healthy RBCs
[159][160][161][162][159,160,161,162]. Virally induced clumping of RBCs, however, is of particular interest for these reasons. First, as noted above, this clumping alone could limit the efficiency of blood oxygenation in the pulmonary capillaries. Second, such clump formation is directly testable both by examination of the blood of COVID-19 patients and by mixing spike protein with RBCs in vitro. Third, as will be detailed, if such virally induced RBC clumping is confirmed, an existing drug that has been found in silico to bind to glycan sites on both spike protein and host cells can be tested in vitro and clinically for inhibition of virally mediated RBC clumping, in conjunction with an anti-COVID-19 therapeutic benefit.
Of the 22 N-linked glycosylation sites at each of the three monomers of SARS-CoV-2 spike protein, eight are located on NTD as noted above, two on RBD, three others elsewhere on S1, and the other nine on S2
[84][85][86][87][84,85,86,87]. N1194 is the closest N-glycosylation site from the C-terminal domain, the end of the spike attached to the virion. In different studies, glycans have been identified as populating between 17 and 21 of these 22 N-glycosylation sites
[86][87][163][164][86,87,163,164]. One study found that ten of these 22 sites have terminal SA moieties, in particular of α5-
N-acetylneuraminic acid (Neu5Ac), the predominant type of SA found in human cells
[25][30][25,30]. The terminal monosaccharides on SARS-CoV-2 spike N-glycans other than SA are galactose, mannose, fucose,
N-acetylglucosamine (GlcNAc) and/or
N-acetylgalactosamine (GalNAc)
[84][87][84,87]. As noted above, two SA-tipped O-linked glycans sites have been identified as well, both on S1 RBD
[86][141][86,141]. Spike protein S1 at its NTD domain was found to bind strongly, in particular to Neu5Ac
[94], the type of SA at SARS-CoV-2 N-glycosylation sites and predominant in human cells.
GPA, the major sialoglycoprotein in human RBCs, is of central interest in the attachment of SARS-CoV-2 to RBCs, as observed in vitro
[35] and on RBCs of COVID-19 patients
[36] as noted above. GPA populates human RBCs at approximately one million molecules per cell and contains most of the SA (of type Neu5Ac) on them
[31][33][74][165][166][31,33,74,165,166]. GPA molecules have the shapes of strands that are anchored approximately 14 nm apart on the RBC plasma membrane, each extending outwards 5 nm
[75]. GPA constitutes the bulk of the RBC’s sialoglycoprotein coating, thus determining its 5 nm thickness
[75][167][168][75,167,168], and accounts for most of its negative charge
[31][169][31,169]. Electrostatic repulsion imposes a minimum distance of approximately 8 nm between the outer boundaries of those sialoglycoprotein coatings of adjoining RBCs in a static suspension, but a smaller separation can be achieved when additional forces are pushing these RBCs together
[168][170][168,170]. SA in its predominant human form, Neu5Ac, is the most common terminal residue of GPA, with its other terminal monosaccharides matching those on SARS-CoV-2 spike N-glycans as noted above: galactose, mannose, fucose,
N-acetylglucosamine (GlcNAc) and
N-acetylgalactosamine (GalNAc)
[74][75][166][74,75,166]. A representation of a portion of an RBC membrane with strands of GPA and with other glycoproteins interspersed is shown in
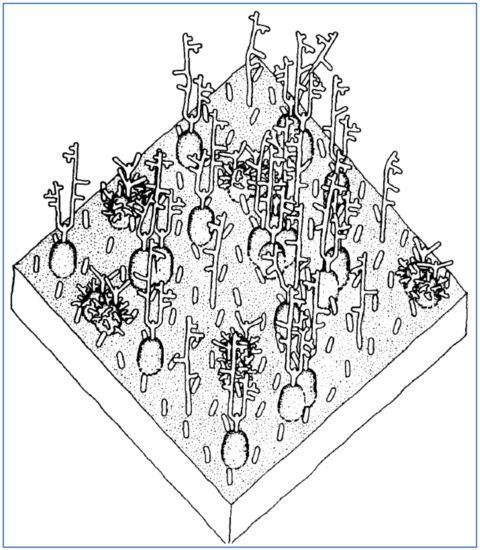
Figure 5.
Figure 5. A representation of a 350 × 350 Angstrom area of the RBC surface depicting its sialoglycoprotein coating, consisting of GPA molecules, extending approximately 5 nm from the RBC cell membrane, plus other smaller glycoprotein molecules interspersed. Reproduced with permission from Elsevier (Viitala, 1985
[75]).