
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Paola Marcato | + 2394 word(s) | 2394 | 2022-03-15 09:06:02 | | | |
| 2 | Nora Tang | Meta information modification | 2394 | 2022-03-25 02:08:25 | | | | |
| 3 | Nora Tang | Meta information modification | 2394 | 2022-03-25 02:10:02 | | |
Video Upload Options
Breast cancer is one of the most common malignancies, with multiple subtypes, based on clinical parameters and molecular profiling. In addition to disease staging, the expression status of hormone receptors’ estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) in tumors define the prognosis of the cancer and treatment options. Hence, clinically, breast cancers are defined as ER+/PR+, HER+, or triple-negative (i.e., those lacking expression of these three receptors). This classification system allows for the administration of endocrine therapies in the hormone expressing subtypes. Additionally, breast cancer subtyping is observed via transcriptome profiling, which has identified four major subtypes (luminal A, luminal B, HER2, and basal-like). The ER+/PR+ breast cancers are predominately luminal A/B and TNBCs are predominately basal-like. Clearly gene expression defines breast cancer; it is then not surprising that the expression of miRNAs also displays subtype-specificity.
1. ER+/PR+ Breast Cancers
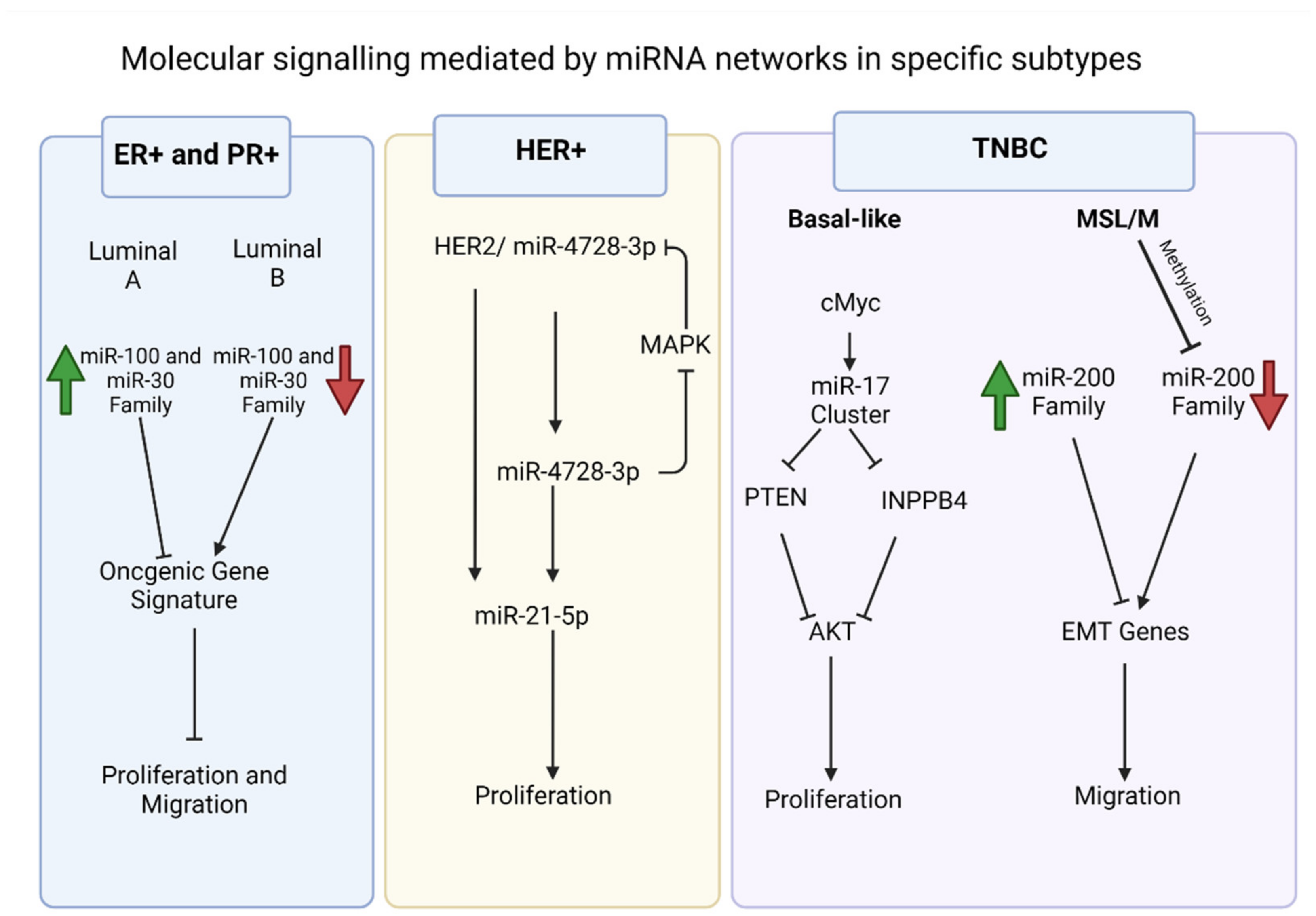
2. HER2 Overexpressing Breast Cancers
3. TNBC
References
- Althoubaity, F.K. Molecular classification of breast cancer: A retrospective cohort study. Ann. Med. Surg. 2019, 49, 44–48.
- Liang, Q.; Ma, D.; Gao, R.-F.; Yu, K.-D. Effect of Ki-67 Expression Levels and Histological Grade on Breast Cancer Early Relapse in Patients with Different Immunohistochemical-based Subtypes. Sci. Rep. 2020, 10, 7648.
- Haakensen, V.D.; Nygaard, V.; Greger, L.; Aure, M.R.; Fromm, B.; Bukholm, I.R.; Lüders, T.; Chin, S.-F.; Git, A.; Caldas, C.; et al. Subtype-specific micro-RNA expression signatures in breast cancer progression. Int. J. Cancer 2016, 139, 1117–1128.
- Endo, Y.; Toyama, T.; Takahashi, S.; Yoshimoto, N.; Iwasa, M.; Asano, T.; Fujii, Y.; Yamashita, H. miR-1290 and its potential targets are associated with characteristics of estrogen receptor α-positive breast cancer. Endocr.-Relat. Cancer 2012, 20, 91–102.
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Taheri, M.; Samadian, M. A Review on the Role of miR-1290 in Cell Proliferation, Apoptosis and Invasion. Front. Mol. Biosci. 2021, 8, 8.
- Amorim, M.; Lobo, J.; Fontes-Sousa, M.; Estevão-Pereira, H.; Salta, S.; Lopes, P.; Coimbra, N.; Antunes, L.; De Sousa, S.P.; Henrique, R.; et al. Predictive and Prognostic Value of Selected MicroRNAs in Luminal Breast Cancer. Front. Genet. 2019, 10, 815.
- Søkilde, R.; Persson, H.; Ehinger, A.; Pirona, A.C.; Fernö, M.; Hegardt, C.; Larsson, C.; Loman, N.; Malmberg, M.; Rydén, L.; et al. Refinement of breast cancer molecular classification by miRNA expression profiles. BMC Genom. 2019, 20, 503.
- McDermott, A.M.; Miller, N.; Wall, D.; Martyn, L.M.; Ball, G.; Sweeney, K.J.; Kerin, M. Identification and Validation of Oncologic miRNA Biomarkers for Luminal A-like Breast Cancer. PLoS ONE 2014, 9, e87032.
- Arabkari, V.; Clancy, E.; Dwyer, R.; Kerin, M.; Kalinina, O.; Holian, E.; Newell, J.; Smith, T.J. Relative and Absolute Expression Analysis of MicroRNAs Associated with Luminal a Breast Cancer—A Comparison. Pathol. Oncol. Res. 2019, 26, 833–844.
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.-H.; Bachelier, R.; Allioli, N.; Hong, S.-S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis—Associated Genes. Cancer Res. 2018, 78, 5259–5273.
- Yin, H.; Wang, Y.; Wu, Y.; Zhang, X.; Zhang, X.; Liu, J.; Wang, T.; Fan, J.; Sun, J.; Yang, A.; et al. EZH2-mediated Epigenetic Silencing of miR-29/miR-30 targets LOXL4 and contributes to Tumorigenesis, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Theranostics 2020, 10, 8494–8512.
- Poudel, S.; Song, J.; Jin, E.-J.; Song, K. Sulfuretin-induced miR-30C selectively downregulates cyclin D1 and D2 and triggers cell death in human cancer cell lines. Biochem. Biophys. Res. Commun. 2013, 431, 572–578.
- Chang, T.-C.; Yu, D.; Lee, Y.-S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50.
- Wang, X.; Li, Y.; Qi, W.; Zhang, N.; Sun, M.; Huo, Q.; Cai, C.; Lv, S.; Yang, Q. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells. Oncotarget 2015, 6, 32737–32747.
- Hu, Y.; Zhu, Q.; Tang, L. MiR-99a Antitumor Activity in Human Breast Cancer Cells through Targeting of mTOR Expression. PLoS ONE 2014, 9, e92099.
- Xia, M.; Li, H.; Wang, J.-J.; Zeng, H.-J.; Wang, S.-H. MiR-99a Suppress Proliferation, Migration and Invasion through Regu-lating Insulin-like Growth Factor 1 Receptor in Breast Cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1755–1763.
- Long, X.; Shi, Y.; Ye, P.; Guo, J.; Zhou, Q.; Tang, Y. MicroRNA-99a Suppresses Breast Cancer Progression by Targeting FGFR3. Front. Oncol. 2020, 9, 1473.
- Iorio, M.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070.
- Estevão-Pereira, H.; Lobo, J.; Salta, S.; Amorim, M.; Lopes, P.; Cantante, M.; Reis, B.; Antunes, L.; Castro, F.; De Sousa, S.P.; et al. Overexpression of circulating MiR-30b-5p identifies advanced breast cancer. J. Transl. Med. 2019, 17, 435.
- Amiruddin, A.; Massi, M.N.; Islam, A.A.; Patellongi, I.; Pratama, M.Y.; Sutandyo, N.; Natzir, R.; Hatta, M.; Latar, N.H.M.; Wahid, S. microRNA-221 and tamoxifen resistance in luminal-subtype breast cancer patients: A case-control study. Ann. Med. Surg. 2021, 73, 103092.
- Loibl, S.; Gianni, L. HER2-positive breast cancer. Lancet 2017, 389, 2415–2429.
- Normann, L.S.; Aure, M.R.; Leivonen, S.-K.; Haugen, M.H.; Hongisto, V.; Kristensen, V.N.; Mælandsmo, G.M.; Sahlberg, K.K. MicroRNA in combination with HER2-targeting drugs reduces breast cancer cell viability in vitro. Sci. Rep. 2021, 11, 10893.
- Baselga, J. Why the Epidermal Growth Factor Receptor? The Rationale for Cancer Therapy. Oncologist 2002, 7 (Suppl. S4), 2–8.
- Baselga, J.; Swain, S.M. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat. Cancer 2009, 9, 463–475.
- Lowery, A.J.; Miller, N.; Devaney, A.; E McNeill, R.; A Davoren, P.; Lemetre, C.; Benes, V.; Schmidt, S.; Blake, J.; Ball, G.; et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neureceptor status in breast cancer. Breast Cancer Res. 2009, 11, R27.
- Luo, Y.; Wang, X.; Niu, W.; Wang, H.; Wen, Q.; Fan, S.; Zhao, R.; Li, Z.; Xiong, W.; Peng, S.; et al. Elevated microRNA-125b levels predict a worse prognosis in HER2-positive breast cancer patients. Oncol. Lett. 2017, 13, 867–874.
- Persson, H.; Kvist, A.; Rego, N.; Staaf, J.; Vallon-Christersson, J.; Luts, L.; Loman, N.; Jönsson, G.B.; Naya, H.; Hoglund, M.; et al. Identification of New MicroRNAs in Paired Normal and Tumor Breast Tissue Suggests a Dual Role for the ERBB2/Her2 Gene. Cancer Res. 2011, 71, 78–86.
- Tashkandi, H.; Shah, N.; Patel, Y.; Chen, H. Identification of new miRNA biomarkers associated with HER2-positive breast cancers. Oncoscience 2015, 2, 924–929.
- Ferracin, M.; Bassi, C.; Pedriali, M.; Pagotto, S.; D’Abundo, L.; Zagatti, B.; Corrà, F.; Musa, G.; Callegari, E.; Lupini, L.; et al. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer 2013, 12, 130.
- Bailey, S.T.; Westerling, T.; Brown, M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2014, 75, 436–445.
- Banzhaf-Strathmann, J.; Edbauer, D. Good guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell Commun. Signal. 2014, 12, 30.
- Newie, I.; Søkilde, R.; Persson, H.; Grabau, D.; Rego, N.; Kvist, A.; Von Stedingk, K.; Axelson, H.; Borg, A.; Vallon-Christersson, J.; et al. The HER2-Encoded miR-4728-3p Regulates ESR1 through a Non-Canonical Internal Seed Interaction. PLoS ONE 2014, 9, e97200.
- Floros, K.V.; Lochmann, T.L.; Hu, B.; Monterrubio, C.; Hughes, M.T.; Wells, J.D.; Morales, C.B.; Ghotra, M.S.; Costa, C.; Souers, A.J.; et al. Coamplification of miR-4728 protects HER2 -amplified breast cancers from targeted therapy. Proc. Natl. Acad. Sci. USA 2018, 115, E2594–E2603.
- Newie, I.; Søkilde, R.; Persson, H.; Jacomasso, T.; Gorbatenko, A.; Borg, Å.; De Hoon, M.; Pedersen, S.F.; Rovira, C. HER2-encoded mir-4728 forms a receptor-independent circuit with miR-21-5p through the non-canonical poly(A) polymerase PAPD5. Sci. Rep. 2016, 6, 35664.
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713.
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18.
- Metzger-Filho, O.; Tutt, A.M.; de Azambuja, E.; Saini, K.S.; Viale, G.; Loi, S.; Bradbury, I.; Bliss, J.; Azim, H.H.A.; Ellis, P.; et al. Dissecting the Heterogeneity of Triple-Negative Breast Cancer. J. Clin. Oncol. 2012, 30, 1879–1887.
- Lü, L.; Mao, X.; Shi, P.; He, B.; Xu, K.; Zhang, S.; Wang, J. MicroRNAs in the Prognosis of Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. Medicine 2017, 96, e7085.
- Gasparini, P.; Cascione, L.; Fassan, M.; Lovat, F.; Guler, G.; Balci, S.; Irkkan, C.; Morrison, C.; Croce, C.M.; Shapiro, C.L.; et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 2014, 5, 1174–1184.
- Du, Y.; Wei, N.; Ma, R.; Jiang, S.; Song, D. A miR-210-3p regulon that controls the Warburg effect by modulating HIF-1α and p53 activity in triple-negative breast cancer. Cell Death Dis. 2020, 11, 731.
- Sanges, F.; Floris, M.; Cossu-Rocca, P.; Muroni, M.R.; Pira, G.; Urru, S.A.M.; Barrocu, R.; Gallus, S.; Bosetti, C.; D’Incalci, M.; et al. Histologic subtyping affecting outcome of triple negative breast cancer: A large Sardinian population-based analysis. BMC Cancer 2020, 20, 491.
- Pareja, F.; Geyer, F.C.; Marchiò, C.; Burke, K.A.; Weigelt, B.; Reis-Filho, J.S. Triple-negative breast cancer: The importance of molecular and histologic subtyping, and recognition of low-grade variants. npj Breast Cancer 2016, 2, 16036.
- De Rinaldis, E.; Gazinska, P.; Mera, A.; Modrusan, Z.; Fedorowicz, G.M.; Burford, B.; Gillett, C.; Marra, P.; Grigoriadis, A.; Dornan, D.; et al. Integrated genomic analysis of triple-negative breast cancers reveals novel microRNAs associated with clinical and molecular phenotypes and sheds light on the pathways they control. BMC Genom. 2013, 14, 643.
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17~92 Family of miRNA Clusters. Cell 2008, 132, 875–886.
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614.
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; E Furth, E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006, 38, 1060–1065.
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843.
- Kalecky, K.; Modisette, R.; Pena, S.; Cho, Y.-R.; Taube, J. Integrative analysis of breast cancer profiles in TCGA by TNBC subgrouping reveals novel microRNA-specific clusters, including miR-17-92a, distinguishing basal-like 1 and basal-like 2 TNBC subtypes. BMC Cancer 2020, 20, 141.
- Liu, H.; Paddock, M.N.; Wang, H.; Murphy, C.J.; Geck, R.C.; Navarro, A.J.; Wulf, G.M.; Elemento, O.; Haucke, V.; Cantley, L.C.; et al. The INPP4B Tumor Suppressor Modulates EGFR Trafficking and Promotes Triple-Negative Breast Cancer. Cancer Discov. 2020, 10, 1226–1239.
- Wang, D.-Y.; Jiang, Z.; Ben-David, Y.; Woodgett, J.R.; Zacksenhaus, E. Molecular stratification within triple-negative breast cancer subtypes. Sci. Rep. 2019, 9, 19107.
- Chakraborty, S.; Krishnan, Y. A structural map of oncomiR-1 at single-nucleotide resolution. Nucleic Acids Res. 2017, 45, 9694–9705.
- Donayo, A.O.; Johnson, R.M.; Tseng, H.-W.; Izreig, S.; Gariepy, A.; Mayya, V.K.; Wu, E.; Alam, R.; Lussier, C.; Jones, R.G.; et al. Oncogenic Biogenesis of pri-miR-17~92 Reveals Hierarchy and Competition among Polycistronic MicroRNAs. Mol. Cell 2019, 75, 340–356.
- Fuziwara, C.S.; Kimura, E.T. Insights into Regulation of the miR-17-92 Cluster of miRNAs in Cancer. Front. Med. 2015, 2, 64.
- Uva, P.; Cossu-Rocca, P.; Loi, F.; Pira, G.; Murgia, L.; Orrù, S.; Floris, M.; Muroni, M.R.; Sanges, F.; Carru, C.; et al. miRNA-135b Contributes to Triple Negative Breast Cancer Molecular Heterogeneity: Different Expression Profile in Basal-like Versus non-Basal-like Phenotypes. Int. J. Med Sci. 2018, 15, 536–548.
- Aakula, A.; Leivonen, S.-K.; Hintsanen, P.; Aittokallio, T.; Ceder, Y.; Børresen-Dale, A.-L.; Perälä, M.; Östling, P.; Kallioniemi, O. MicroRNA-135b regulates ERα, AR and HIF1AN and affects breast and prostate cancer cell growth. Mol. Oncol. 2015, 9, 1287–1300.
- Hua, K.; Jin, J.; Zhao, J.; Song, J.; Song, H.; Li, D.; Maskey, N.; Zhao, B.; Wu, C.; Xu, H.; et al. miR-135b, upregulated in breast cancer, promotes cell growth and disrupts the cell cycle by regulating LATS2. Int. J. Oncol. 2016, 48, 1997–2006.
- Lv, Z.; Xin, H.; Yang, Z.; Wang, W.; Dong, J.; Jin, L.; Li, F. miR-135b promotes proliferation and metastasis by targeting APC in triple-negative breast cancer. J. Cell. Physiol. 2019, 234, 10819–10826.
- Pu, T.; Shen, M.; Li, S.; Yang, L.; Gao, H.; Xiao, L.; Zhong, X.; Zheng, H.; Liu, Y.; Ye, F.; et al. Repression of miR-135b-5p promotes metastasis of early-stage breast cancer by regulating downstream target SDCBP. Lab. Investig. 2019, 99, 1296–1308.





