Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shih-Hwa Chiou | + 1792 word(s) | 1792 | 2022-02-14 07:47:08 | | | |
| 2 | Dean Liu | -4 word(s) | 1788 | 2022-03-10 08:09:18 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Chiou, S. Circular RNAs Modulate Cancer Hallmark and Molecular Pathways. Encyclopedia. Available online: https://encyclopedia.pub/entry/20410 (accessed on 05 March 2026).
Chiou S. Circular RNAs Modulate Cancer Hallmark and Molecular Pathways. Encyclopedia. Available at: https://encyclopedia.pub/entry/20410. Accessed March 05, 2026.
Chiou, Shih-Hwa. "Circular RNAs Modulate Cancer Hallmark and Molecular Pathways" Encyclopedia, https://encyclopedia.pub/entry/20410 (accessed March 05, 2026).
Chiou, S. (2022, March 10). Circular RNAs Modulate Cancer Hallmark and Molecular Pathways. In Encyclopedia. https://encyclopedia.pub/entry/20410
Chiou, Shih-Hwa. "Circular RNAs Modulate Cancer Hallmark and Molecular Pathways." Encyclopedia. Web. 10 March, 2022.
Copy Citation
Circular RNAs (circRNAs) are noncoding products of backsplicing of pre-mRNAs which have been established to possess potent biological functions. Due to their circular nature, the are characterized by high stability. Dysregulated circRNA expression has been linked to diseases including different types of cancer.
circular RNA
cancer progression
metastasis
1. Circular RNAs (CircRNAs): Discovery, Biogenesis, and Degradation
1.1. Circular RNA Discovery
Originally, circular RNAs (circRNAs) were identified in eukaryotic cells in 1979 [1]. CircRNAs were initially considered as transcription noise devoid of any functionality. The advent of next-generation sequencing (NGS) in the early 2000s revealed the importance of noncoding transcriptome. CircRNAs were identified as noncoding RNA species with increased stability due to their circular nature, which fostered considerable investigation of their roles in the biological processes and disease [2][3].
1.2. CircRNA Biogenesis and Properties
CircRNAs are now known to be transcribed from protein-coding genes and further processed by unconventional pre-mRNA splicing mechanism, referred to as backsplicing, in which the 3′-end of an exon is ligated to the 5′-end donor splice site of the same or an upstream exon [3][4]. The conventional spliceosome and canonical splicing sites are essential for the process of backsplicing. Indeed, the process of backsplicing competes with conventional linear splicing [5]. Based on their structure derived from different modes of biogenesis, circRNAs can be classified as exonic (ecircRNAs), exon-intronic (EIciRNAs), circularized intronic (ciRNAs), and tRNA intronic (tricRNAs), with the former constituting 85% of all circRNAs. In the process of circRNA biogenesis, the distant donor and acceptor splice sites are selectively brought into proximity to be ligated, and three major models accounting for it have been delineated:
-
In a lariat-driven circularization, the exon-skipping linear splicing event produces a lariat that includes the skipped exons. Such lariat structure can further undergo internal splicing generating EIciRNAs or ecircRNAs (Figure 1A).
-
In intron pairing-driven circularization, also known as direct backsplicing, the splice sites are brought to close proximity by complementary pairing between inverted repeats (e.g., Alu repeats) flanking circularized exons (Figure 1B).
-
In RNA-binding protein (RBP)-driven circularization, the splice sites in intronic sequences are bridged together by certain trans-factor RBPs (Figure 1C).
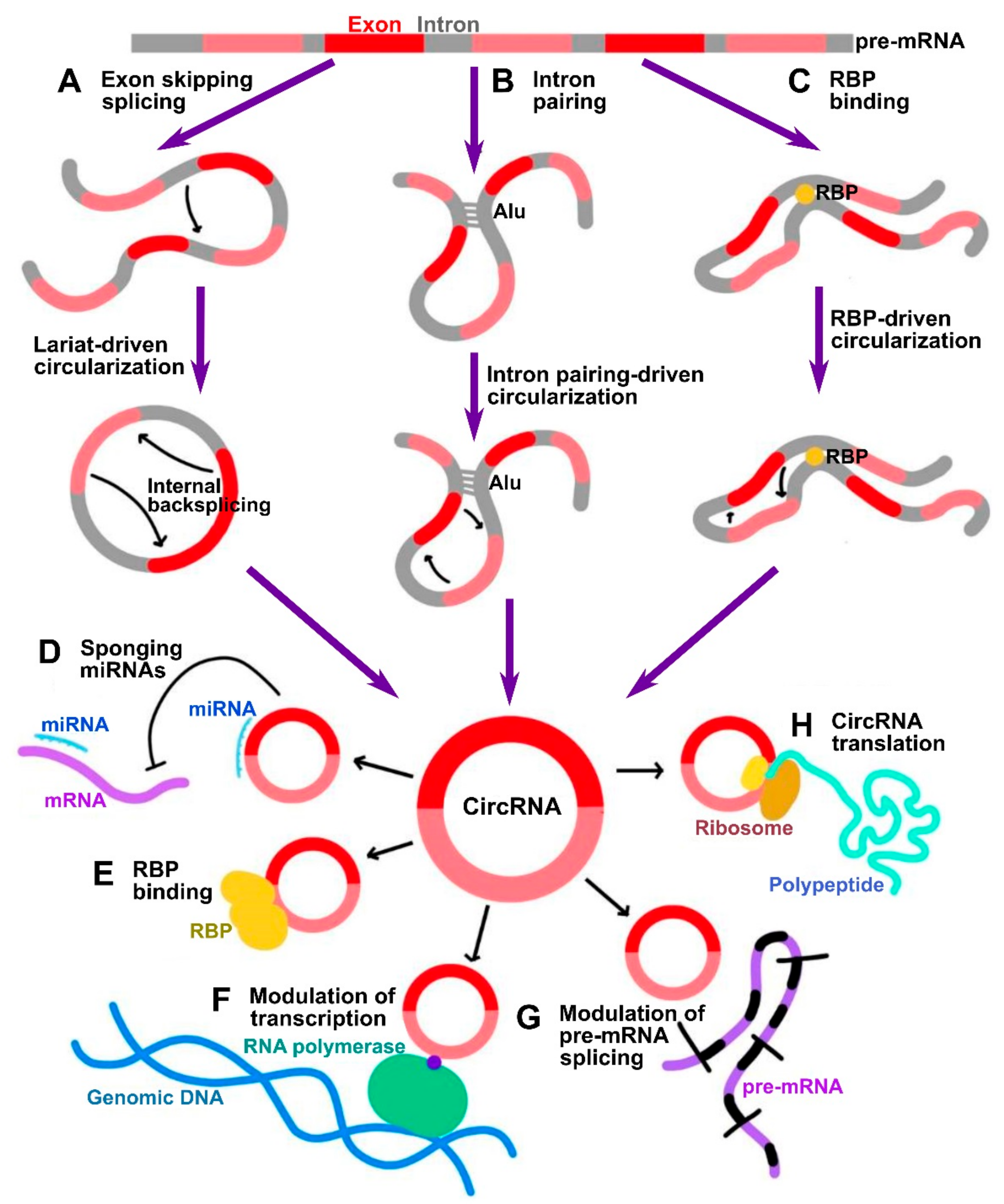
Figure 1. Summary of circRNA biogenesis and functional mechanisms. (A–C) Three major mechanisms of circRNA biogenesis by which distant backspliced sites are brought into proximity: (A) in lariat-driven circularization, circRNA is generated by internal backsplicing within a lariat formed by exon skipping direct splicing; (B) in intron pairing-driven circularization, backspliced sites are brought into proximity by pairing between inverted repeats such as Alu; (C) in RBP binding-driven circularization, backsplicing is facilitated by RBPs. (D–H) Summary of major biological functions of circRNAs: (D) miRNA sponging; (E) modulation of functions of RBPs; (F) modulation of transcription; (G) modulation of splicing; (H) translation of peptides.
Given the lower efficiency of backsplicing as compared to linear splicing, the production rate of circRNAs is lower than that of their linear counterparts. However, the lack of free ends renders them resistant to the exonuclease-mediated degradation which results in longer half-life and higher number within cells [6]. Based on this property, circRNAs can be used to serve as novel diagnostics markers or disease targets. In 2019, it was first reported that circRNAs undergo biodegradation by RNase L global RNA-degrading endonuclease [7]. In such a mechanism, circRNAs were found to be stabilized by binding to inactive unphosphorylated protein kinase R (PKR) that blocked circRNAs’ accessibility to RNase L [7][8]. Phosphorylation of PKR activates it and mediates its dissociation from circRNAs, which, in turn, exposes them to active RNase L [7] (Figure 2). Similarly, another study showed that the highly structured nature of circRNAs ensures their degradation via binding to RNA-binding protein UPF1 and its associated protein G3BP1, which causes subsequent degradation by RNase P or RNase MRP [9]. Therefore, researchers can conclude that the cell achieves circRNA homeostasis by possessing a well-controlled circRNA biogenesis and degradation systems (Figure 2). Dysregulation of these processes can lead to cellular aberrations and disease. Numerous transcriptome analysis studies identified more than 20,000 circRNAs expressed from 60% of total genes in eukaryote cells. Intriguingly, a portion of circRNAs is expressed in a tissue- or developmental stage-specific manner indicative of temporal and spatial control of their expression. CircRNAs have been identified in numerous species by different high-throughput RNA deep sequencing projects. In one of the studies, it was identified that around 14.4% of human fibroblast circRNAs can be linked to 69 murine testis orthologous circRNAs [10]. Such orthologous conservation features of circRNA imply the importance of their role in gene regulation.
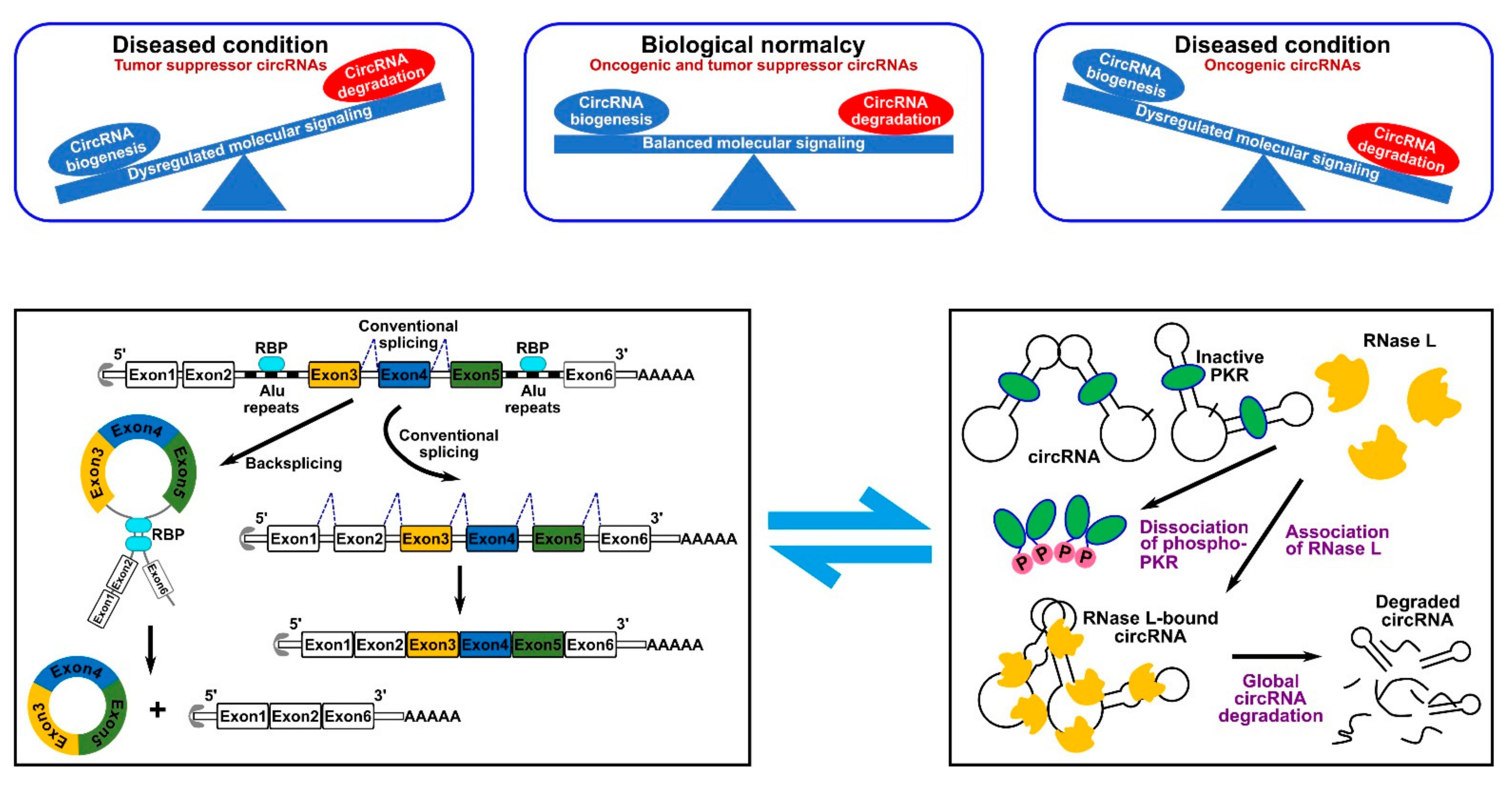
Figure 2. CircRNA homeostasis determines normal biological conditions or disease state. (Top panel) such homeostasis is determined by the fine balance between circRNA biogenesis and degradation mechanisms. (Bottom panel) schematic representation of a typical RBP-driven biogenesis mechanism (left) and of the RNase L-dependent degradation mechanism (right).
2. Molecular Functions of CircRNAs
CircRNAs have been proven to be involved in various biological processes and mechanistically have been widely characterized to conduct their functions by several major mechanisms:
-
One of the most investigated functions of circRNAs is their ability to indirectly control gene expression through sponging of microRNAs (miRNAs) [6] (Figure 1D). CircRNAs often contain miRNA response elements (MREs) which can bind miRNAs, thus preventing them from silencing their gene targets [11][12]. By such mechanism, these circRNAs are classified as competing endogenous RNAs (ceRNAs) [11]. Among the most prominent of them is ciRS-7 (CDR1as), which was discovered to possess up to 70 target sites for miR-7 [6]. Therefore, a vast number of studies has been focused on investigating ceRNA mode of action of circRNAs. However, large-scale screening studies indicate that usually circRNAs contain a limited number of MREs, indicative that this is not the only bona fide mechanism.
-
CircRNAs can associate with RNA binding proteins (RBPs) and thus mediate different aspects of their functionality (Figure 1E). For instance, circRNAs can act as scaffolds facilitating the formation of protein complexes. For example, circ-FOXO3 was shown to interact with both MDM2 and p53, which promoted MDM2-dependent ubiquitination and degradation of p53 [13]. The same circRNA was also shown to form ternary complex with CDK2 and p21, preventing the association of the former into functional complexes with cyclins A and E, thus acting by decoy mechanism [14].
-
CircRNAs that retain the intronic sequences of their parental genes, such as ciRNAs and EIciRNAs, are predominantly localized in the nucleus. As was shown by Pol II CLIP, EIciRNAs such as circEIF3J and circPAIP2 could associate with RNA polymerase II and enhance their parental gene expression in cis in a U1 snRNP-dependent manner [15] (Figure 1F). The broad-scale effects of circRNAs on pre-mRNA splicing are not widely characterized as yet; however, it has been known that circularization and linear splicing compete with each other on a co-transcriptional level (Figure 1G). Such regulatory mechanism was demonstrated for MBL/MBNL1 gene encoding muscleblind (MBL) splicing factor. The protein product of this gene was shown to directly promote circularization of the exon 2 of its host gene by direct binding to the flanking sequences in the adjacent introns to produce circMBL, concomitantly reducing linear splicing [5]. Finally, in addition to regulating mRNAs at transcriptional and splicing levels, circRNAs were also shown to regulate the stability of mRNAs. For example, the degraded fragments of CDR1, as circRNA could downregulate CDR1 by promoting degradation via antisense pairing [16]. On the contrary, circRasGEF1B could enhance the stability of its target ICAM1 mRNA [17].
-
Although circRNAs have traditionally been regarded as noncoding RNAs given the lack of the 5′ cap that is normally required for the initiation of translation, the discovery of internal ribosome entry sites (IRES) within multiple circRNAs suggests that they may be extensively translated into peptides or proteins in 5′ cap-independent manner [18] (Figure 1H). One prominent example of such circRNAs is circ-ZNF609, whose IRES-dependent translation was shown to play an important role in myogenesis [19]. In addition, it was shown that initiation of translation in circRNAs can be driven by m6A methylation. CircRNAs were found to be enriched in consensus m6A motifs, whose methylation by METTL3/14 methyl transferase complex promoted the translation, and demethylation by FTO demethylase inhibited the translation [20]. In such a mechanism, m6A deposited on circRNAs could be recognized by YTHDF3 m6A reader protein, which in turn recruited eIF4G2 initiation factor [20].
3. CircRNA as Therapeutics Molecular Pathway Targets in Cancer
RNA therapeutic technology provides the opportunity of targeting every coding and noncoding gene in biological systems. Interestingly, RNA therapeutics efficacy has been proven to be workable by few approved RNA drugs for liver, muscle, and rare genetic diseases [21]. Considering that many circRNAs were confirmed to promote cancer progression, targeting them offers a great potential therapeutic strategy for combating various cancers [22].
Overall, circRNA studies focus more on circRNA differential expression in cancers based on the premises that circRNAs can serve as more reliable and accessible liquid biomarkers for cancer detection [23]. Other than the diagnostic value, targeting circRNA-mediated molecular pathways for therapeutic purposes is also conceivable if the full molecular mechanisms of action of the most dysregulated circRNAs implicated in cancers are known. Currently, this perspective still remains unrealized in clinical practice [24]. Therefore, gain-of-function and loss-of-function strategies are deployed in circRNA research in order to harness dysregulated circRNAs for cancer therapy [24]. Precisely, tumor suppressor circRNAs are ectopically expressed in order to replenish the downregulated tumor suppressor circRNAs in cancers [25]. For example, several transient circRNA overexpression DNA vectors harboring the Alu repeats that facilitate circularization have been developed [4][26][27]. Likewise, lentivirus and AAV delivery systems have also been used to generate stable circRNA ectopic expression in vitro and in vivo [28].
In contrast, oncogenic circRNAs are normally targeted for degradation using several techniques. For instance, RNA interference (RNAi) methods represent the most prominent circRNA degradation strategy currently available. Researchers have used both the conventional short interference RNA (siRNA) [29][30] and the more lasting short hairpin RNA (shRNA) techniques to degrade copious oncogenic circRNAs [31][32].
In addition to siRNAs and shRNAs, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) systems represent an effective and promising approach for targeting circRNAs. Whereas CRISPR/Cas9 is normally used to edit DNA [33], CRISPR/Cas13 is a recently discovered system that has been utilized to target RNA [34]. At present, both systems have been used to modulate the expressions of circRNAs [25][35]. In one of the studies, CRISPR/Cas9 was used to delete the complementary Alu repeat in HIPK3 gene, thus preventing the expression of circHIPK3 [36]. Similarly, circRNA CDR1as was also downregulated using CRISPR/Cas9 and gRNA targeting CDR1as locus [37]. This evidence proved the possibility of therapeutic targeting of circRNAs using the CRISPR/Cas9 system; however, the concerns are raised about affecting the linear counterparts of circRNAs which can lead to off-target effects [24]. Therefore, various CRISPR/Cas13 orthologs have been investigated for circRNA targeting. CRISPR/Cas13d was used to downregulate oncogenic circFAM120A [24]. Similarly, circZKSCAN1 was also effectively downregulated using CRISPR/Cas13d system [38]. More detailed information on CRISPR/Cas system-mediated targeting of circRNAs can be found in recent reviews [24][25].
References
- Hsu, M.-T.; Coca-Prados, M.J.N. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340.
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98.
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.J.N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338.
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E.J.G. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015, 29, 2168–2182.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J.J.N. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.J.C. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 2019, 177, 865–880.e21.
- Guo, Y.; Wei, X.; Peng, Y.J.T. Structure-mediated degradation of CircRNAs. Trends Cell Biol. 2020, 30, 501–503.
- Fischer, J.W.; Busa, V.F.; Shao, Y.; Leung, A.K.L. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell 2020, 78, 70–84.e6.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E.J.R. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157.
- Ala, U.J.C. Competing endogenous RNAs, non-coding RNAs and diseases: An intertwined story. Cells 2020, 9, 1574.
- Barrett, S.P.; Salzman, J.J.D. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847.
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370.
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.J.N.s.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J.J.T.E.j. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422.
- Ng, W.L.; Marinov, G.K.; Liau, E.S.; Lam, Y.L.; Lim, Y.-Y.; Ea, C.-K. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016, 13, 861–871.
- Chen, C.K.; Cheng, R.; Demeter, J.; Chen, J.; Weingarten-Gabbay, S.; Jiang, L.; Snyder, M.P.; Weissman, J.S.; Segal, E.; Jackson, P.K.; et al. Structured elements drive extensive circular RNA translation. Mol. Cell 2021, 81, 4300–4318.e13.
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37.e9.
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.J.C.r. Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Res. 2017, 27, 626–641.
- Yu, A.-M.; Choi, Y.H.; Tu, M.-J.J.P.R. RNA drugs and RNA targets for small molecules: Principles, progress, and challenges. Pharmacol. Rev. 2020, 72, 862–898.
- Floor, S.L.; Dumont, J.E.; Maenhaut, C.; Raspe, E. Hallmarks of cancer: Of all cancer cells, all the time? Trends Mol Med. 2012, 18, 509–515.
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90.
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185.
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Circular RNAs as Therapeutic Agents and Targets. Front. Physiol. 2018, 9, 1262.
- Pan, H.; Li, T.; Jiang, Y.; Pan, C.; Ding, Y.; Huang, Z.; Yu, H.; Kong, D. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J. Cell Biochem. 2018, 119, 440–446.
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.-L.; Cherry, S.; Wilusz, J.E. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell 2017, 68, 940–954.e943.
- Meganck, R.M.; Liu, J.; Hale, A.E.; Simon, K.E.; Fanous, M.M.; Vincent, H.A.; Wilusz, J.E.; Moorman, N.J.; Marzluff, W.F.; Asokan, A. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 2021, 23, 821–834.
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S.J.C.d.; et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018, 9, 417.
- Li, Y.; Zheng, F.; Xiao, X.; Xie, F.; Tao, D.; Huang, C.; Liu, D.; Wang, M.; Wang, L.; Zeng, F.; et al. Circ HIPK 3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017, 18, 1646–1659.
- Pamudurti, N.R.; Patop, I.L.; Krishnamoorthy, A.; Ashwal-Fluss, R.; Bartok, O.; Kadener, S. An in vivo strategy for knockdown of circular RNAs. Cell Discov. 2020, 6, 52.
- Li, L.; Li, W.; Chen, N.; Zhao, H.; Xu, G.; Zhao, Y.; Pan, X.; Zhang, X.; Zhou, L.; Yu, D.; et al. FLI1 Exonic Circular RNAs as a Novel Oncogenic Driver to Promote Tumor Metastasis in Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 1302–1317.
- Ablain, J.; Zon, L. Tissue-specific gene targeting using CRISPR/Cas9. Methods Cell Biol. 2016, 135, 189–202.
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280–284.
- Li, S.; Li, X.; Xue, W.; Zhang, L.; Yang, L.Z.; Cao, S.M.; Lei, Y.N.; Liu, C.X.; Guo, S.K.; Shan, L.; et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 2021, 18, 51–59.
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215.
- Yang, X.; Li, S.; Wu, Y.; Ge, F.; Chen, Y.; Xiong, Q. The circular RNA CDR1as regulate cell proliferation via TMED2 and TMED10. BMC Cancer 2020, 20, 312.
- Zhang, Y.; Nguyen, T.M.; Zhang, X.O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021, 22, 41.
More
Information
Subjects:
Cell Biology; Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
536
Revisions:
2 times
(View History)
Update Date:
10 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





