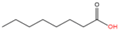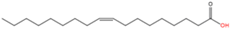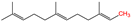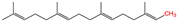| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shenglong Zhu | + 2852 word(s) | 2852 | 2022-03-03 03:03:58 | | | |
| 2 | Peter Tang | Meta information modification | 2852 | 2022-03-04 03:32:06 | | |
Video Upload Options
Proteins play indispensable roles in maintaining cell survival, and their functions are often regulated by post-translational modifications (PTMs), in which proteins are proteolytically cleaved or enzymatically conjugated with modifying groups. Various enzymes, including kinases, phosphatases, transferases, and ligases, catalyze approximately 500 discrete PTMs of a diverse set of proteins. PTMs regulate diverse activities of a colossal number of proteins. For example, various types of lipids can be covalently linked to proteins enzymatically or non-enzymatically. Protein lipidation is perhaps not as extensively studied as protein phosphorylation, ubiquitination, or glycosylation although it is no less significant than these modifications. Evidence suggests that proteins can be attached by at least seven types of lipids, including fatty acids, lipoic acids, isoprenoids, sterols, phospholipids, glycosylphosphatidylinositol anchors, and lipid-derived electrophiles.
1. Introduction
2. Types of Protein Lipidation
2.1. Fatty Acylation
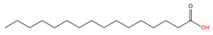
2.1.1. S-palmitoylation
|
Modification |
Lipid |
Structure |
Linkage |
Modified Residue |
References |
|
|---|---|---|---|---|---|---|
|
1 |
S-palmitoylation |
Palmitic acid (C16:0) |
|
Thioester |
Cysteine |
|
|
2 |
N-terminal palmitoylation |
Palmitic acid (C16:0) |
Amide |
N-terminal Cysteine |
||
|
3 |
Nε-palmitoylation |
Palmitic acid (C16:0) |
Amide |
Lysine |
||
|
4 |
O-palmitoylation |
Palmitic acid (C16:0) |
Oxyester |
Serine |
[47] |
|
|
Threonine |
[48] |
|||||
|
5 |
N-terminal myristoylation |
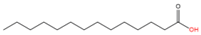
Myristic acid (C14:0) |
|
Amide |
N-terminal Glycine |
[49] |
|
6 |
Nε-myristoylation |
Myristic acid (C14:0) |
Amide |
Lysine |
||
|
7 |
S-stearoylation |
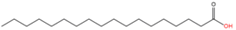
Stearic acid (C18:0) |
|
Thioester |
Cysteine |
|
|
8 |
O-octanoylation |
Octanoic acid (C8:0) |
|
Oxyester |
Serine |
|
|
9 |
O-palmitoleoylation |
Palmitoleic acid (C16:1n7) |
|
Oxyester |
Serine |
|
|
10 |
N-oleoylation |
Oleic acid (C18:1n9) |
|
Amide |
Lysine |
[60] |
|
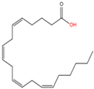
11 |
Unnamed |
Arachidonic acid (C20:4n6) |
|
Yet unknown |
Yet unknown |
[61] |
|
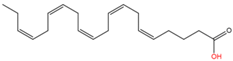
12 |
Unnamed |
Eicosapentaenoic acid (C20:5n3) |
|
Yet unknown |
Yet unknown |
[61] |
|
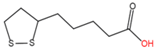
13 |
N-lipoylation |
Lipoic acid |
|
Amide |
Lysine |
|
|
14 |
S-prenylation |
Isoprenoid |
|
Untitled |
C-terminal Cysteine |
|
|
|
||||||
|
15 |
C-terminal phosphatidyl-ethanolaminylation |
PE |
|
Amide |
C-terminal Glycine |
|
|
16 |
C-terminal cholesterolyation |
Cholesterol |
|
Oxyester |
C-terminus |
|
|
17 |
C-terminal GPI anchor |
GPI |
|
Amide |
C-terminus |
|
|
18 |
LDE acylation |
LDE |
|
Carbonyls |
Nucleophilic residues |
|
|
Aldehydes |
||||||
N-system nomenclature was used for the fatty acids (the order of carbon atoms starts from the methyl carbon of the fatty acid).
2.1.2. N-palmitoylation
2.1.3. O-palmitoylation
2.1.4. N-myristoylation
2.1.5. Acylation of Other Saturated Fatty Acids
2.1.6. Acylation of Unsaturated Fatty Acids
2.2. N-lipoylation
2.3. S-prenylation
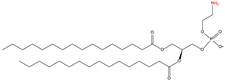
2.4. C-terminal Phosphatidylethanolaminylation
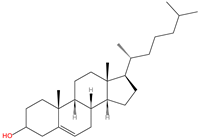
2.5. C-terminal Cholesterolyation
2.6. C-terminal GPI Anchoring
2.7. LDE Acylation
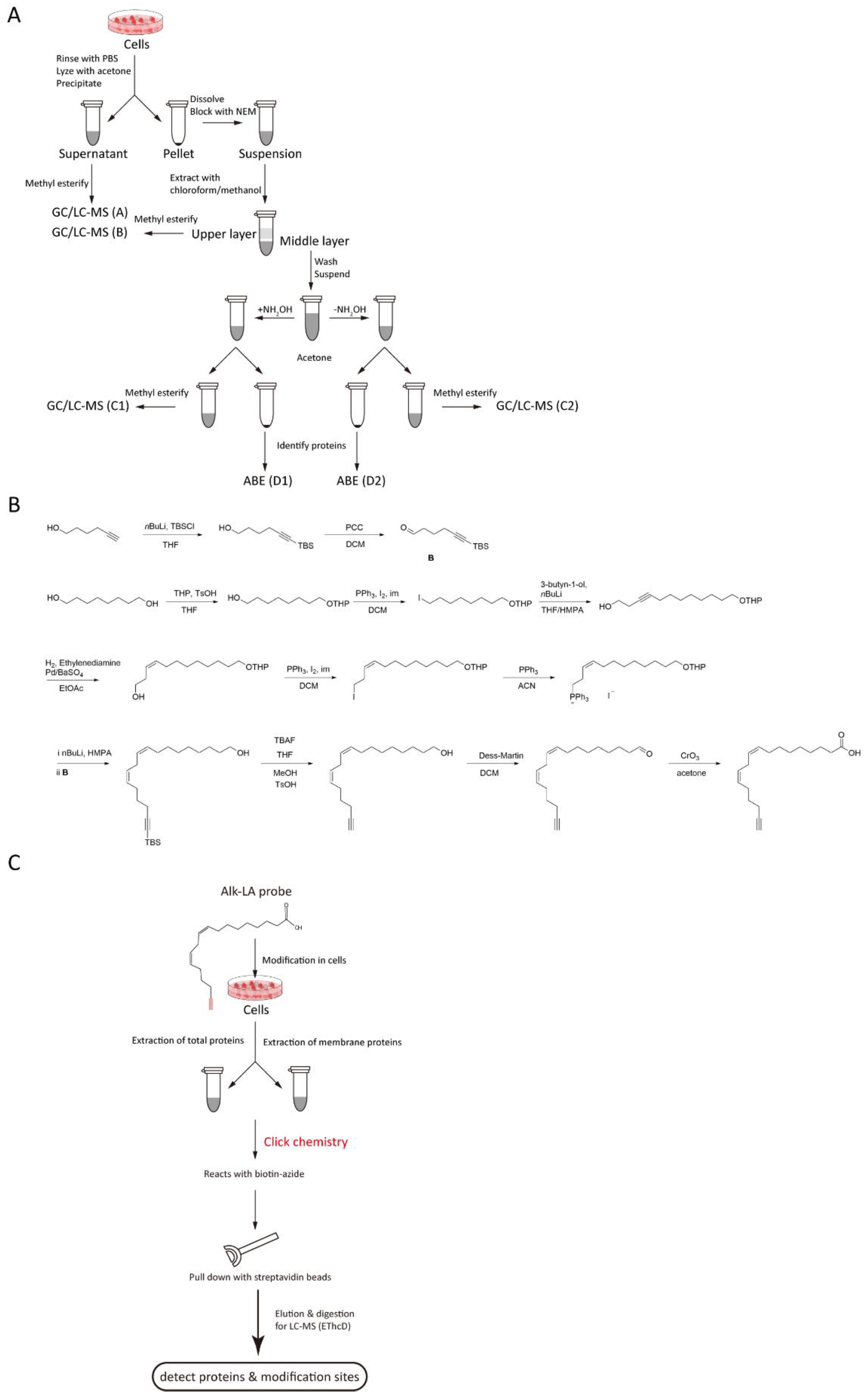
3. Detection of Protein Lipidation
4. Detection of PUFA-Modified Proteins
4.1. Difficulties in Detecting PUFA-Modified Proteins
4.2. Limitations in Current Methodology
4.3. Potential Solutions
References
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261.
- Keenan, E.K.; Zachman, D.K.; Hirschey, M.D. Discovering the landscape of protein modifications. Mol. Cell 2021, 81, 1868–1878.
- Zavialova, M.G.; Zgoda, V.G.; Nikolaev, E.N. Analysis of contribution of protein phosphorylation in the development of the diseases. Biomeditsinskaia Khimiia 2017, 63, 101–114.
- Cifani, P.; Kentsis, A. Towards comprehensive and quantitative proteomics for diagnosis and therapy of human disease. Proteomics 2017, 17, 1600079.
- Yang, M.; Zhang, Y.; Ren, J. Acetylation in cardiovascular diseases: Molecular mechanisms and clinical implications. Biochim. et Biophys. Acta (BBA)—Mol. Basis Dis. 2020, 1866, 165836.
- Torres, M.P.; Dewhurst, H.; Sundararaman, N. Proteome-wide Structural Analysis of PTM Hotspots Reveals Regulatory Elements Predicted to Impact Biological Function and Disease. Mol. Cell. Proteom. 2016, 15, 3513–3528.
- Chen, B.; Sun, Y.; Niu, J.; Jarugumilli, G.K.; Wu, X. Protein Lipidation in Cell Signaling and Diseases: Function, Regulation, and Therapeutic Opportunities. Cell Chem. Biol. 2018, 25, 817–831.
- Seo, J.; Lee, K.J. Post-translational modifications and their biological functions: Proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 2004, 37, 35–44.
- Fukata, Y.; Fukata, M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 2010, 11, 161–175.
- Greaves, J.; Chamberlain, L.H. New links between S-acylation and cancer. J. Pathol. 2014, 233, 4–6.
- Yeste-Velasco, M.; Linder, M.E.; Lu, Y.J. Protein S-palmitoylation and cancer. Biochim. Biophys. Acta Rev. Cancer 2015, 1856, 107–120.
- Chavda, B.; Arnott, J.A.; Planey, S.L. Targeting protein palmitoylation: Selective inhibitors and implications in disease. Expert Opin. Drug Discov. 2014, 9, 1005–1019.
- Roth, A.F.; Wan, J.; Bailey, A.O.; Sun, B.; Kuchar, J.A.; Green, W.N.; Phinney, B.S.; Yates, J.R., 3rd; Davis, N.G. Global analysis of protein palmitoylation in yeast. Cell 2006, 125, 1003–1013.
- Peng, T.; Thinon, E.; Hang, H.C. Proteomic analysis of fatty-acylated proteins. Curr. Opin. Chem. Biol. 2016, 30, 77–86.
- Hannoush, R.N.; Sun, J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat. Chem. Biol. 2010, 6, 498–506.
- Mueller, T.M.; Meador-Woodruff, J.H. Post-translational protein modifications in schizophrenia. NPJ Schizophr. 2020, 6, 5.
- Hong, M.; Zhang, Y.; Hu, F. Membrane protein structure and dynamics from NMR spectroscopy. Annu. Rev. Phys. Chem. 2012, 63, 1–24.
- Miles, A.J.; Wallace, B.A. Circular dichroism spectroscopy of membrane proteins. Chem. Soc. Rev. 2016, 45, 4859–4872.
- Hanashima, S.; Nakane, T.; Mizohata, E. Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins. Membranes 2021, 11, 823.
- Clabbers, M.T.B.; Xu, H. Macromolecular crystallography using microcrystal electron diffraction. Acta Crystallogr. D Struct. Biol. 2021, 77, 313–324.
- Aldini, G.; Domingues, M.R.; Spickett, C.M.; Domingues, P.; Altomare, A.; Sanchez-Gomez, F.J.; Oeste, C.L.; Perez-Sala, D. Protein lipoxidation: Detection strategies and challenges. Redox Biol. 2015, 5, 253–266.
- Linder, M.E.; Deschenes, R.J. Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007, 8, 74–84.
- Resh, M.D. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006, 2, 584–590.
- Braun, P.E.; Radin, N.S. Interactions of lipids with a membrane structural protein from myelin. Biochemistry 1969, 8, 4310–4318.
- Schmidt, M.F.; Schlesinger, M.J. Fatty acid binding to vesicular stomatitis virus glycoprotein: A new type of post-translational modification of the viral glycoprotein. Cell 1979, 17, 813–819.
- Schlesinger, M.J.; Magee, A.I.; Schmidt, M.F. Fatty acid acylation of proteins in cultured cells. J. Biol. Chem. 1980, 255, 10021–10024.
- Stoffyn, P.; Folch-Pi, J. On the type of linkage binding fatty acids present in brain white matter proteolipid apoprotein. Biochem. Biophys. Res. Commun. 1971, 44, 157–161.
- Chen, B.; Zheng, B.; DeRan, M.; Jarugumilli, G.K.; Fu, J.; Brooks, Y.S.; Wu, X. ZDHHC7-mediated S-palmitoylation of Scribble regulates cell polarity. Nat. Chem. Biol. 2016, 12, 686–693.
- Hernandez, J.L.; Davda, D.; Cheung See Kit, M.; Majmudar, J.D.; Won, S.J.; Gang, M.; Pasupuleti, S.C.; Choi, A.I.; Bartkowiak, C.M.; Martin, B.R. APT2 Inhibition Restores Scribble Localization and S-Palmitoylation in Snail-Transformed Cells. Cell Chem. Biol. 2017, 24, 87–97.
- Chan, P.; Han, X.; Zheng, B.; DeRan, M.; Yu, J.; Jarugumilli, G.K.; Deng, H.; Pan, D.; Luo, X.; Wu, X. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat. Chem. Biol. 2016, 12, 282–289.
- Lanyon-Hogg, T.; Faronato, M.; Serwa, R.A.; Tate, E.W. Dynamic Protein Acylation: New Substrates, Mechanisms, and Drug Targets. Trends Biochem. Sci. 2017, 42, 566–581.
- Blanc, M.; David, F.; Abrami, L.; Migliozzi, D.; Armand, F.; Burgi, J.; van der Goot, F.G. SwissPalm: Protein Palmitoylation database. F1000Research 2015, 4, 261.
- Blanc, M.; David, F.P.A.; van der Goot, F.G. SwissPalm 2: Protein S-Palmitoylation Database. Methods Mol. Biol. 2019, 2009, 203–214.
- Dowal, L.; Yang, W.; Freeman, M.R.; Steen, H.; Flaumenhaft, R. Proteomic analysis of palmitoylated platelet proteins. Blood 2011, 118, e62–e73.
- Kang, R.; Wan, J.; Arstikaitis, P.; Takahashi, H.; Huang, K.; Bailey, A.O.; Thompson, J.X.; Roth, A.F.; Drisdel, R.C.; Mastro, R.; et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 2008, 456, 904–909.
- Martin, B.R.; Wang, C.; Adibekian, A.; Tully, S.E.; Cravatt, B.F. Global profiling of dynamic protein palmitoylation. Nat Methods 2011, 9, 84–89.
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The Palmitoylation Machinery Is a Spatially Organizing System for Peripheral Membrane Proteins. Cell 2010, 141, 458–471.
- Adachi, N.; Hess, D.T.; McLaughlin, P.; Stamler, J.S. S-Palmitoylation of a Novel Site in the beta2-Adrenergic Receptor Associated with a Novel Intracellular Itinerary. J. Biol. Chem. 2016, 291, 20232–20246.
- Rossin, A.; Durivault, J.; Chakhtoura-Feghali, T.; Lounnas, N.; Gagnoux-Palacios, L.; Hueber, A.O. Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability. Cell Death Differ. 2015, 22, 643–653.
- Frohlich, M.; Dejanovic, B.; Kashkar, H.; Schwarz, G.; Nussberger, S. S-palmitoylation represents a novel mechanism regulating the mitochondrial targeting of BAX and initiation of apoptosis. Cell Death Dis. 2014, 5, e1057.
- Fredericks, G.J.; Hoffmann, F.W.; Rose, A.H.; Osterheld, H.J.; Hess, F.M.; Mercier, F.; Hoffmann, P.R. Stable expression and function of the inositol 1,4,5-triphosphate receptor requires palmitoylation by a DHHC6/selenoprotein K complex. Proc. Natl. Acad. Sci. USA 2014, 111, 16478–16483.
- Aramsangtienchai, P.; Spiegelman, N.A.; Cao, J.; Lin, H.N. S-Palmitoylation of Junctional Adhesion Molecule C Regulates Its Tight Junction Localization and Cell Migration. J. Biol. Chem. 2017, 292, 5325–5334.
- Cai, H.; Smith, D.A.; Memarzadeh, S.; Lowell, C.A.; Cooper, J.A.; Witte, O.N. Differential transformation capacity of Src family kinases during the initiation of prostate cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 6579–6584.
- Varland, S.; Osberg, C.; Arnesen, T. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 2015, 15, 2385–2401.
- Madsen, A.S.; Andersen, C.; Daoud, M.; Anderson, K.A.; Laursen, J.S.; Chakladar, S.; Huynh, F.K.; Colaco, A.R.; Backos, D.S.; Fristrup, P.; et al. Investigating the Sensitivity of NAD+-dependent Sirtuin Deacylation Activities to NADH. J. Biol. Chem. 2016, 291, 7128–7141.
- Zhang, X.; Spiegelman, N.A.; Nelson, O.D.; Jing, H.; Lin, H. SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. eLife 2017, 6, 39.
- Zou, C.; Ellis, B.M.; Smith, R.M.; Chen, B.B.; Zhao, Y.; Mallampalli, R.K. Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J. Biol. Chem. 2011, 286, 28019–28025.
- Branton, W.D.; Rudnick, M.S.; Zhou, Y.; Eccleston, E.D.; Fields, G.B.; Bowers, L.D. Fatty acylated toxin structure. Nature 1993, 365, 496–497.
- Maurer-Stroh, S.; Eisenhaber, B.; Eisenhaber, F. N-terminal N-myristoylation of proteins: Prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 2002, 317, 541–557.
- Liu, Z.; Yang, T.; Li, X.; Peng, T.; Hang, H.C.; Li, X.D. Integrative chemical biology approaches for identification and characterization of “erasers” for fatty-acid-acylated lysine residues within proteins. Angew. Chem. Int. Ed. Engl. 2015, 54, 1149–1152.
- Teng, Y.B.; Jing, H.; Aramsangtienchai, P.; He, B.; Khan, S.; Hu, J.; Lin, H.; Hao, Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci. Rep. 2015, 5, 8529.
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550.
- Brett, K.; Kordyukova, L.V.; Serebryakova, M.V.; Mintaev, R.R.; Alexeevski, A.V.; Veit, M. Site-specific S-acylation of influenza virus hemagglutinin: The location of the acylation site relative to the membrane border is the decisive factor for attachment of stearate. J. Biol. Chem. 2014, 289, 34978–34989.
- Kordyukova, L.V.; Serebryakova, M.V.; Baratova, L.A.; Veit, M. S acylation of the hemagglutinin of influenza viruses: Mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 2008, 82, 9288–9292.
- Gutierrez, J.A.; Solenberg, P.J.; Perkins, D.R.; Willency, J.A.; Knierman, M.D.; Jin, Z.; Witcher, D.R.; Luo, S.; Onyia, J.E.; Hale, J.E. Ghrelin octanoylation mediated by an orphan lipid transferase. Proc. Natl. Acad. Sci. USA 2008, 105, 6320–6325.
- Yang, J.; Brown, M.S.; Liang, G.; Grishin, N.V.; Goldstein, J.L. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell 2008, 132, 387–396.
- Takada, R.; Satomi, Y.; Kurata, T.; Ueno, N.; Norioka, S.; Kondoh, H.; Takao, T.; Takada, S. Monounsaturated fatty acid modification of Wnt protein: Its role in Wnt secretion. Dev. Cell 2006, 11, 791–801.
- Zhai, L.; Chaturvedi, D.; Cumberledge, S. Drosophila wnt-1 undergoes a hydrophobic modification and is targeted to lipid rafts, a process that requires porcupine. J. Biol. Chem. 2004, 279, 33220–33227.
- Kakugawa, S.; Langton, P.F.; Zebisch, M.; Howell, S.; Chang, T.H.; Liu, Y.; Feizi, T.; Bineva, G.; O—Reilly, N.; Snijders, A.P.; et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature 2015, 519, 187–192.
- Schey, K.L.; Gutierrez, D.B.; Wang, Z.; Wei, J.; Grey, A.C. Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry 2010, 49, 9858–9865.
- Muszbek, L.; Laposata, M. Covalent modification of proteins by arachidonate and eicosapentaenoate in platelets. J. Biol. Chem. 1993, 268, 18243–18248.
- Mathias, R.A.; Greco, T.M.; Oberstein, A.; Budayeva, H.G.; Chakrabarti, R.; Rowland, E.A.; Kang, Y.B.; Shenk, T.; Cristea, I.M. Sirtuin 4 Is a Lipoamidase Regulating Pyruvate Dehydrogenase Complex Activity. Cell 2014, 159, 1615–1625.
- Rowland, E.A.; Greco, T.M.; Snowden, C.K.; McCabe, A.L.; Silhavy, T.J.; Cristea, I.M. Sirtuin Lipoamidase Activity Is Conserved in Bacteria as a Regulator of Metabolic Enzyme Complexes. Mbio 2017, 8, e01096-17.
- Moores, S.L.; Schaber, M.D.; Mosser, S.D.; Rands, E.; Ohara, M.B.; Garsky, V.M.; Marshall, M.S.; Pompliano, D.L.; Gibbs, J.B. Sequence Dependence of Protein Isoprenylation. J. Biol. Chem. 1991, 266, 14603–14610.
- Zhang, F.L.; Casey, P.J. Protein prenylation: Molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996, 65, 241–269.
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178.
- Ray, A.; Jatana, N.; Thukral, L. Lipidated proteins: Spotlight on protein-membrane binding interfaces. Prog. Biophys. Mol. Biol. 2017, 128, 74–84.
- Chen, M.H.; Li, Y.J.; Kawakami, T.; Xu, S.M.; Chuang, P.T. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004, 18, 641–659.
- Goetz, J.A.; Singh, S.; Suber, L.M.; Kull, F.J.; Robbins, D.J. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J. Biol. Chem. 2006, 281, 4087–4093.
- Yu, S.C.; Guo, Z.W.; Johnson, C.; Gu, G.F.; Wu, Q.Y. Recent progress in synthetic and biological studies of GPI anchors and GPI-anchored proteins. Curr. Opin. Chem. Biol. 2013, 17, 1006–1013.
- Masuishi, Y.; Nomura, A.; Okayama, A.; Kimura, Y.; Arakawa, N.; Hirano, H. Mass spectrometric identification of glycosylphosphatidylinositol-anchored peptides. J. Proteome Res. 2013, 12, 4617–4626.
- Chen, Y.; Qin, W.; Wang, C. Chemoproteomic profiling of protein modifications by lipid-derived electrophiles. Curr. Opin. Chem. Biol. 2016, 30, 37–45.
- Nagahara, N.; Matsumura, T.; Okamoto, R.; Kajihara, Y. Protein cysteine modifications: (1) medical chemistry for proteomics. Curr. Med. Chem. 2009, 16, 4419–4444.
- George, J.; Soares, C.; Montersino, A.; Beique, J.C.; Thomas, G.M. Palmitoylation of LIM Kinase-1 ensures spine-specific actin polymerization and morphological plasticity. eLife 2015, 4, e06327.
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 1998, 273, 14037–14045.
- Buglino, J.A.; Resh, M.D. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 2008, 283, 22076–22088.
- Suzuki, T.; Moriya, K.; Nagatoshi, K.; Ota, Y.; Ezure, T.; Ando, E.; Tsunasawa, S.; Utsumi, T. Strategy for comprehensive identification of human N-myristoylated proteins using an insect cell-free protein synthesis system. Proteomics 2010, 10, 1780–1793.
- Patwardhan, P.; Resh, M.D. Myristoylation and membrane binding regulate c-Src stability and kinase activity. Mol. Cell Biol. 2010, 30, 4094–4107.
- Thinon, E.; Serwa, R.A.; Broncel, M.; Brannigan, J.A.; Brassat, U.; Wright, M.H.; Heal, W.P.; Wilkinson, A.J.; Mann, D.J.; Tate, E.W. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 2014, 5, 4919.
- Liang, J.; Xu, Z.X.; Ding, Z.; Lu, Y.; Yu, Q.; Werle, K.D.; Zhou, G.; Park, Y.Y.; Peng, G.; Gambello, M.J.; et al. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat. Commun. 2015, 6, 7926.
- Stackpole, E.E.; Akins, M.R.; Fallon, J.R. N-myristoylation regulates the axonal distribution of the Fragile X-related protein FXR2P. Mol. Cell. Neurosci. 2014, 62, 42–50.
- Kumar, S.; Parameswaran, S.; Sharma, R.K. Novel myristoylation of the sperm-specific hexokinase 1 isoform regulates its atypical localization. Biol. Open 2015, 4, 1679–1687.
- Wright, M.H.; Heal, W.P.; Mann, D.J.; Tate, E.W. Protein myristoylation in health and disease. J. Chem. Biol. 2010, 3, 19–35.
- Chen, Y.; Chen, W.; Cobb, M.H.; Zhao, Y.M. PTMap-A sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc. Natl. Acad. Sci. USA 2009, 106, 761–766.
- Huang, H.; Sabari, B.R.; Garcia, B.A.; Allis, C.D.; Zhao, Y. SnapShot: Histone modifications. Cell 2014, 159, 458–458.e1.
- DeMar, J.C., Jr.; Anderson, R.E. Identification and quantitation of the fatty acids composing the CoA ester pool of bovine retina, heart, and liver. J. Biol. Chem. 1997, 272, 31362–31368.
- Liang, X.; Nazarian, A.; Erdjument-Bromage, H.; Bornmann, W.; Tempst, P.; Resh, M.D. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J. Biol. Chem. 2001, 276, 30987–30994.
- Dizhoor, A.M.; Ericsson, L.H.; Johnson, R.S.; Kumar, S.; Olshevskaya, E.; Zozulya, S.; Neubert, T.A.; Stryer, L.; Hurley, J.B.; Walsh, K.A. The Nh2 Terminus of Retinal Recoverin Is Acylated by a Small Family of Fatty-Acids. J. Biol. Chem. 1992, 267, 16033–16036.
- Kokame, K.; Fukada, Y.; Yoshizawa, T.; Takao, T.; Shimonishi, Y. Lipid Modification at the N-Terminus of Photoreceptor G-Protein Alpha-Subunit. Nature 1992, 359, 749–752.
- Pereira-Leal, J.B.; Hume, A.N.; Seabra, M.C. Prenylation of Rab GTPases: Molecular mechanisms and involvement in genetic disease. FEBS Lett. 2001, 498, 197–200.
- Rowland, E.A.; Snowden, C.K.; Cristea, I.M. Protein lipoylation: An evolutionarily conserved metabolic regulator of health and disease. Curr. Opin. Chem. Biol. 2018, 42, 76–85.
- Reed, L.J. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J. Biol. Chem. 2001, 276, 38329–38336.
- Jiang, H.; Zhang, X.; Chen, X.; Aramsangtienchai, P.; Tong, Z.; Lin, H. Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies. Chem. Rev. 2018, 118, 919–988.
- Liu, M.; Sjogren, A.K.; Karlsson, C.; Ibrahim, M.X.; Andersson, K.M.; Olofsson, F.J.; Wahlstrom, A.M.; Dalin, M.; Yu, H.; Chen, Z.; et al. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 6471–6476.
- Sjogren, A.K.M.; Andersson, K.M.E.; Khan, O.; Olofsson, F.J.; Karlsson, C.; Bergo, M.O. Inactivating GGTase-I reduces disease phenotypes in a mouse model of K-RAS-induced myeloproliferative disease. Leukemia 2011, 25, 186–189.
- Porter, J.A.; Ekker, S.C.; Park, W.J.; von Kessler, D.P.; Young, K.E.; Chen, C.H.; Ma, Y.; Woods, A.S.; Cotter, R.J.; Koonin, E.V.; et al. Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 1996, 86, 21–34.
- Zeng, X.; Goetz, J.A.; Suber, L.M.; Scott, W.J.; Schreiner, C.M.; Robbins, D.J. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature 2001, 411, 716–720.
- Orlean, P.; Menon, A.K. Thematic review series: Lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011.
- Tsai, Y.H.; Liu, X.; Seeberger, P.H. Chemical biology of glycosylphosphatidylinositol anchors. Angew. Chem. Int. Ed. Engl. 2012, 51, 11438–11456.
- Gamage, D.G.; Hendrickson, T.L. GPI transamidase and GPI anchored proteins: Oncogenes and biomarkers for cancer. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 446–464.
- Bautista, J.M.; Marin-Garcia, P.; Diez, A.; Azcarate, I.G.; Puyet, A. Malaria proteomics: Insights into the parasite-host interactions in the pathogenic space. J. Proteom. 2014, 97, 107–125.
- Puig, B.; Altmeppen, H.; Glatzel, M. The GPI-anchoring of PrP: Implications in sorting and pathogenesis. Prion 2014, 8, 11–18.
- Wang, C.; Weerapana, E.; Blewett, M.M.; Cravatt, B.F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 2014, 11, 79–85.
- Boveris, A.; Navarro, A. Brain mitochondrial dysfunction in aging. IUBMB Life 2008, 60, 308–314.
- Chen, Y.; Liu, Y.; Hou, X.; Ye, Z.; Wang, C. Quantitative and Site-Specific Chemoproteomic Profiling of Targets of Acrolein. Chem. Res. Toxicol. 2019, 32, 467–473.
- Galligan, J.J.; Rose, K.L.; Beavers, W.N.; Hill, S.; Tallman, K.A.; Tansey, W.P.; Marnett, L.J. Stable histone adduction by 4-oxo-2-nonenal: A potential link between oxidative stress and epigenetics. J. Am. Chem. Soc. 2014, 136, 11864–11866.
- Cui, Y.W.; Li, X.; Lin, J.W.; Hao, Q.; Li, X.D. Histone Ketoamide Adduction by 4-Oxo-2-nonenal Is a Reversible Posttranslational Modification Regulated by Sirt2. ACS Chem. Biol. 2017, 12, 47–51.
- Bantscheff, M.; Lemeer, S.; Savitski, M.M.; Kuster, B. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012, 404, 939–965.
- Bantscheff, M.; Schirle, M.; Sweetman, G.; Rick, J.; Kuster, B. Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 2007, 389, 1017–1031.
- Held, J.M.; Gibson, B.W. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol. Cell Proteom. 2012, 11, R111.013037.
- Hao, G.; Derakhshan, B.; Shi, L.; Campagne, F.; Gross, S.S. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA 2006, 103, 1012–1017.
- Bolla, J.R.; Agasid, M.T.; Mehmood, S.; Robinson, C.V. Membrane Protein-Lipid Interactions Probed Using Mass Spectrometry. Annu. Rev. Biochem. 2019, 88, 85–111.
- Urner, L.H.; Schulze, M.; Maier, Y.B.; Hoffmann, W.; Warnke, S.; Liko, I.; Folmert, K.; Manz, C.; Robinson, C.V.; Haag, R.; et al. A new azobenzene-based design strategy for detergents in membrane protein research. Chem. Sci. 2020, 11, 3538–3546.
- Morgner, N.; Montenegro, F.; Barrera, N.P.; Robinson, C.V. Mass spectrometry—From peripheral proteins to membrane motors. J. Mol. Biol. 2012, 423, 1–13.
- Kim, M.S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581.
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587.
- Fuentes, N.R.; Kim, E.; Fan, Y.Y.; Chapkin, R.S. Omega-3 fatty acids, membrane remodeling and cancer prevention. Mol. Asp. Med. 2018, 64, 79–91.
- Yang, W.S.; Kim, K.J.; Gaschler, M.M.; Patel, M.; Shchepinov, M.S.; Stockwell, B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. USA 2016, 113, E4966–E4975.
- Shaikh, S.R.; Edidin, M. Polyunsaturated fatty acids, membrane organization, T cells, and antigen presentation. Am. J. Clin. Nutr. 2006, 84, 1277–1289.
- Shaikh, S.R.; Kinnun, J.J.; Leng, X.; Williams, J.A.; Wassall, S.R. How polyunsaturated fatty acids modify molecular organization in membranes: Insight from NMR studies of model systems. Biochim. Biophys. Acta 2015, 1848, 211–219.
- Cravatt, B.F.; Simon, G.M.; Yates, J.R. 3rd, The biological impact of mass-spectrometry-based proteomics. Nature 2007, 450, 991–1000.
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207.
- Domon, B.; Aebersold, R. Mass spectrometry and protein analysis. Science 2006, 312, 212–217.
- Sorek, N.; Akerman, A.; Yalovsky, S. Analysis of protein prenylation and S-acylation using gas chromatography-coupled mass spectrometry. Methods Mol. Biol. 2013, 1043, 121–134.
- Zhou, B.; Wang, Y.; Yan, Y.; Mariscal, J.; Di Vizio, D.; Freeman, M.R.; Yang, W. Low-Background Acyl-Biotinyl Exchange Largely Eliminates the Coisolation of Non-S-Acylated Proteins and Enables Deep S-Acylproteomic Analysis. Anal. Chem. 2019, 91, 9858–9866.
- Schulte-Zweckel, J.; Dwivedi, M.; Brockmeyer, A.; Janning, P.; Winter, R.; Triola, G. A hydroxylamine probe for profiling S-acylated fatty acids on proteins. Chem. Commun. 2019, 55, 11183–11186.
- Windsor, K.; Genaro-Mattos, T.C.; Kim, H.Y.; Liu, W.; Tallman, K.A.; Miyamoto, S.; Korade, Z.; Porter, N.A. Probing lipid-protein adduction with alkynyl surrogates: Application to Smith-Lemli-Opitz syndrome. J. Lipid Res. 2013, 54, 2842–2850.
- Zheng, B.; Jarugumilli, G.K.; Chen, B.; Wu, X. Chemical Probes to Directly Profile Palmitoleoylation of Proteins. Chembiochem 2016, 17, 2022–2027.
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Pasa-Tolic, L. High-throughput proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454.
- Pan, J.; Borchers, C.H. Top-down mass spectrometry and hydrogen/deuterium exchange for comprehensive structural characterization of interferons: Implications for biosimilars. Proteomics 2014, 14, 1249–1258.
- Zinnel, N.F.; Pai, P.J.; Russell, D.H. Ion mobility-mass spectrometry (IM-MS) for top-down proteomics: Increased dynamic range affords increased sequence coverage. Anal. Chem. 2012, 84, 3390–3397.