Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hueng-Chuen Fan | + 2429 word(s) | 2429 | 2022-01-28 09:51:16 | | | |
| 2 | Catherine Yang | Meta information modification | 2429 | 2022-03-03 10:25:14 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Fan, H. Etiology and Pathogenesis of Glioblastoma Multiforme. Encyclopedia. Available online: https://encyclopedia.pub/entry/20134 (accessed on 07 February 2026).
Fan H. Etiology and Pathogenesis of Glioblastoma Multiforme. Encyclopedia. Available at: https://encyclopedia.pub/entry/20134. Accessed February 07, 2026.
Fan, Hueng-Chuen. "Etiology and Pathogenesis of Glioblastoma Multiforme" Encyclopedia, https://encyclopedia.pub/entry/20134 (accessed February 07, 2026).
Fan, H. (2022, March 03). Etiology and Pathogenesis of Glioblastoma Multiforme. In Encyclopedia. https://encyclopedia.pub/entry/20134
Fan, Hueng-Chuen. "Etiology and Pathogenesis of Glioblastoma Multiforme." Encyclopedia. Web. 03 March, 2022.
Copy Citation
Glioblastoma multiforme (GBM) is a type of brain tumor that is notorious for its aggressiveness and invasiveness, and the complete removal of GBM is still not possible, even with advanced diagnostic strategies and extensive therapeutic plans. Its dismal prognosis and short survival time after diagnosis make it a crucial public health issue.
glioblastoma multiforme (GBM)
1. Isocitrate Dehydrogenase (IDH)
To elucidate genetic alterations in GBM patients with varying prognoses or responses to specific targeted therapies and to identify subgroups of GBM patients for a better histopathological classification, an integrated genomic analysis was used to identify mutations inisocitrate dehydrogenase 1(IDH1) in 12% of patients with GBM [1]. It was subsequently reported that GBMs without IDH1 mutations often have mutations of isocitrate dehydrogenase 2 (IDH2) [2]. The structures of IDH1 (located on chromosome 2q33.3) and IDH2 (located on chromosome 15q26.1) are homodimeric and share similar sequences and an almost identical protein structure [3]. IDH1 and IDH2 encode two separate, different isocitrate dehydrogenase enzymes, which are nicotinamide adenine dinucleotide phosphate (NADP+)-dependent, catalyze oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG), and reduce oxidized nicotinamide adenine dinucleotide (NAD+) or NADP+ to NADH or NADPH. IDH1, localizing in the cytoplasm and peroxisomes, involves cellular metabolism and protection from reactive oxygen species and radiation [4][5]. IDH2, localizing in the mitochondria, is associated with the regulation of the tricarboxylic acid cycle and protection from oxidative stress [5]. DH1 and IDH2 mutations are mono-allelic, somatic, and missense changes. Mutations in IDH1 commonly affect R132, which is the binding site for isocitrate [1]. Mutations in IDH2 only affect R172 and R140 [2][6]. Mutant IDH possesses catalytic activity to convert α-KG to 2-hydroxyglutarate (2-HG) [7]. Excessive 2-HG is a metabolic hallmark of certain subtypes of gliomas [8]. Both mutated IDH1 and IDH2 are common in adult gliomas (WHO grades II and III) and secondary GBM (WHO grade IV). Mutated IDH1 and IDH2 are very rare in childhood GBM [9], suggesting gliomas with mutated IDH are clinically and genetically different from those with wild-type (WT) IDH genes.
2. Co-Deletion at Chromosome Regions 1p/19q
The complete deletion of chromosomes 1p and 19q is common in oligodendrogliomas and occur in 50–70% of both low-grade and anaplastic tumors [10][11][12][13]. These findings suggest that chromosomes 1p and 19q may contain tumor suppressor genes, including the far upstream element binding protein 1 (FUBP1) on chromosome 1p and the capicua transcriptional repressor (CIC) on chromosome 19q [14][15][16]. The FUBP1 expresses a single-stranded DNA-binding protein that can bind to several DNA regions, which harbor the far upstream element (FUSE) that is localized in the upstream of c-Myc. One function of FUSE is to regulate c-Myc in undifferentiated cells [17]. CIC, a tissue-specific transcriptional repressor, is expressed in the developing CNS and its dysfunction is associated with spinocerebellar ataxia type 1. This CIC-DNA interaction can be inhibited through the activation of the receptor tyrosine kinase (RTK) core signaling molecule mitogen-activated protein kinase (MAPK), which then allows for the transcription of CIC targets through this RTK-MAPK signaling axis. CIC alterations are common in specific cancer types (e.g., oligodendroglioma and Ewing-like sarcomas) [18]. Two clinical trials have clarified associations between combined 1p/19q co-deletion and an improved chemotherapeutic response and prognosis in oligodendrogliomas [19][20]. However, partial 1p or 19q deletion is more common in astrocytic tumors and secondary GBM but rare in primary GBM [21][22][23].
3. Mutations of H3F3A
Inside the nucleus, DNA, RNA and proteins form chromatin, which packs the DNA to a smaller volume and prevents the long DNA strands from being tangled. The structure of the chromatin is like “beads on a string”. The nucleosome is the “bead”, which is a basic unit of chromatin. Histones, nuclear proteins, can store DNA, modulate chromatin structure, impact gene expression, and regulate almost all DNA metabolic processes through post-translational modification, which includes methylation, phosphorylation, acetylation, ubiquitylation, and sumoylation [24]. Each nucleosome is composed of a histone octamer, which is composed of two copies each of the core histone H2A, H2B, H3, and H4. H1, which does not contribute to the nucleosome bead, binds to the linker DNA region between nucleosomes, helping pack the chromatin into higher order structures. A chain of nucleosomes is compacted to form a chromosome [25]. Histone H3s, which have several variants, include H3.1, H3.2, and H3.3. H3.1 is encoded by HIST1H3A-J.H3.2 is encoded by HIST2H3A, HIST2H3C, and HIST2H3D. H3.3 is encoded by H3F3A and H3F3B [26]. Numerous H3 lysine residues can be post-translationally modified, including acetylated at lysines 9, 14, 18, 23, and 56; methylated at arginine 2 and lysines 4, 9, 27, 36, and 79, and phosphorylated at ser10, ser28, Thr3, and Thr11 [27]. Mutation of H3F3A, including K27M substitution, in which a lysine residue on the histone H3 tail is substituted for a methionine, were discovered in diffused intrinsic pontine glioma(DIPG) [28]. As the location of the H3F3A at lysine 27 is at or near critical regulatory histone residues, therefore, the alternations of mutant K27M on genes, which should be silent, produce widespread aberrant DNA methylation and deregulation of gene expression, impede physiological differentiation and to drive cell transformation [29]. Furthermore, in the WHO CNS tumor classification 2016, the principle of an integrated diagnosis was introduced with the combination of histological and molecular features, exemplified in the novel entity “diffuse midline glioma, H3K27M- mutant” [30]. The mutation in HIST1H3B-C, have been detected in approximately 10% of DIPG [31]. Another mutation of H3F3A, encoding a glycine 34 to arginine or valine (G34R/V) substitution, is reported in a smaller portion of pediatric and young adult high-grade astrocytoma [9]. H3F3G34 mutations may drive pediatric GBM through mismatch repair (MMR) deficiency [32] and upregulation of v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) (MYCN) [33].
4. Telomeres and Telomerase
A telomere is a capping structure located at the end of a linear chromosome. As most prokaryotes have circular chromosomes, they do not have telomeres; on the other hand, telomeres in vertebrates consist of a region of repeats of the six-nucleotide sequence TTAGGG at the ends of chromosomes, which themselves carry the complementary DNA strand sequence, AATCCC. In humans, the TTAGGG sequence repeats in tandem approximately 3000–20,000 times [34][35]. The sequence is bound by the shelterin complex, which is formed by TRF1, TRF2, TPP1, TIN2, POT1 and RAP1 [36]. Telomeres are known to maintain the stability of chromosomes and protect genes [37][38]. Chromosomes without this capping structure, meanwhile, become truncated and fuse with neighboring chromosomes [39]. In the process of chromosome replication, the synthesis of Okazaki fragments requires that RNA primers attach to the lagging strand. The shedding RNA then causes telomere shortening. As such, the accumulative loss of telomeres can eventually cause cell cycle arrest and apoptosis, leading to speculation that the progressive reduction in telomere length may play a key role in determining the timing of in vitro cellular senescence [40][41]. Therefore, telomeres are also viewed by some as a sort of biological clock that controls normal cell proliferation [42]. It is estimated that the human telomere loses approximately 24.8 to 27.7 base pairs per year [43][44]. There are several factors, however, that affect the rate of telomere shortening, including the host’s age [45]; gender [46]; genetic and epigenetic regulation [47][48][49]; social and economic background [50][51], and life style factors, such as the lack or presence of exercise, obesity, smoking, and unhealthy diets [44][51][52] (Figure 1). Individuals with shorter telomeres are known to be associated with various age-related diseases and conditions, such as heart failure [53], coronary heart disease [54][55][56], diabetes [57], osteoporosis [58], and a shorter lifespan [59][60][61]. Although the shorter length of telomeres is generally thought to be a marker of poor health and aging, it can lead to genomic instability [62][63][64] and elevated telomerase activity (TA) [65][66], resulting in a potential cancer predisposition factor.
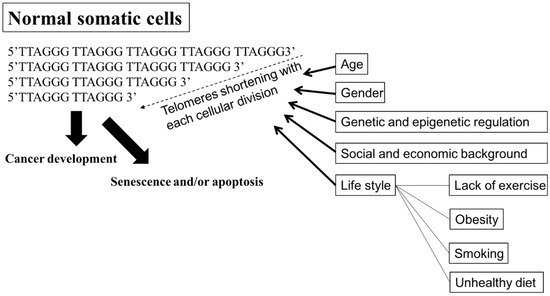
Figure 1. Factors affecting the rate of shortening telomeres. Telomeres are regions of repetitive nucleotide sequences, TTAGGG. Telomeres shorten in each cell division due to incomplete DNA replication. When telomere length progressively reduces to a critical point, the cell then executes senescence or apoptosis, or develops cancer. Several factors, including age; gender; genetic and epigenetic regulation; social and economic background; and life style, such as lack of exercise, obesity, smoking, and an unhealthy diet, are associated with increasing the rate of losing telomeres, leading to senescence or apoptosis, premature death, and cancer development. Bold arrow: shortening DNA leading to senescence and/or apoptosis, or cancer development. Arrow: factors affecting telomeres shortening.
Cell division leads to progressive telomere shortening, resulting in cell senescence; however, the shortening of telomeres can be counteracted by telomerase [67]. Telomerase, an RNA-dependent DNA polymerase, is expressed in developing embryos, in reproductive cells (i.e., proliferating germlines), in activated immune cells, and transiently, in adult stem cells, but telomerase is turned off in most adult human tissues. In immortalized cells, telomere length remains stable, with the activation of telomerase being considered one of the main mechanisms underlying this stability [67][68][69]. TA is exhibited in almost all human tumors and in tumor-derived cell lines, while most human somatic cells do not display TA, except for highly proliferative cells, such as bone marrow cells [70][71]. Telomerase, which is made up of TERT, TERC, and specialized proteins (e.g., dyskerin), preserves telomere stability by adding TTAGGG repeats to the end of the given chromosome (capping) in rapidly dividing cells [68][69][72], using its complementary TERC sequence as the template [73], together with TERT subunit as the catalytic component [67]. Activities of TERT are frequently up-regulated in human cancers, which is thought to be a critical mechanism contributing to human tumorigenesis [74][75]. Studies have identified two cancer-specific TERT promoter mutations (C228T and C250T) [76] in the activation of telomerase in various cancer cells [77][78], including GBM [79]. The two mutations in the TERT, which cause TERT activation to increase TA to elongate telomere length [76][80], lead to the proliferative, anti-senescence, and immortal properties of tumor cells. Mutations in the TERT promoter have been detected in more than 50% of primary adult GBM, and these mutations are correlated with EGFR, IDH1, IDH2, TP53, and ATX mutations and increased TA [81][80][82][83][84]. Mutations in the TERT promoter have been detected in 3–7% of pediatric GBM [76][80] and tumor cells in this group maintain or increase telomere length through alternative lengthening of telomeres (ALT) pathway [85][86].
5. Alternative Lengthening of Telomeres, α-Thalassemia X-Linkedintellectual Disability, and Death-Domain-Associated Protein
TA was detected in 76% of cervical cancer cases [87], 54% of medullary thyroid cancer cases [88], 42% of well-differentiated papillary thyroid cancer cases [89], 86.6% of non-small cell lung cancer cases [90], and > 80% of hepatocellular carcinoma cases [91]. Although telomerase reactivation is the most common mechanism of telomeric repeat addition in cancers [65], there is a convincing argument that TA is not the only way for tumors to become immortalized; otherwise, these tumors would have shrunk and died unless they managed to boost their TA to maintain their telomere length. Additionally, Indeed, 5–10% cancer cases exploit a telomerase-independent mechanism to elongate their telomeres, a phenomenon that is also known as ALT [92]. In addition to immortalized cells and cancer cells, ALT exists in non-neoplastic tissues, in endothelial, stromal, and some epithelial cells [93]. Although the prevalence of the ALT phenotype in cancers is low, ALT is common in certain cancer subtypes, including gliomas [94]. While primary GBM has been found to employ telomerase activation, nearly 75% of WHO grades II–III astrocytomas and secondary GBMs, with normal telomerase expression and WT TERT promoter, was observed to employ ALT for the maintenance of telomere length and genome stability [15][95]. The transformation from telomerase-dependent to ALT-mediated telomere lengthening is considered one of the strategies cancer cells adopt to escape cell senescence and apoptosis caused by telomerase dysfunction or absence [96][97]. ALT, which is not present in normal cells, often begins with a loss of chromatin remodeling proteins in the telomeres, with a resulting DNA damage response, recombination, and abnormal protein behavior that initiates ALT [98].
α-thalassemia X-linked intellectual disability (ATRX) syndrome is characterized by distinctive craniofacial features, genital anomalies, severe developmental delays, hypotonia, intellectual disability, and mild-to-moderate anemia secondary to α-thalassemia [99].The ATRX gene, which encodes a SWItch (SWI)/sucrose non-fermenting(SNF)-like chromatin remodeling protein, is frequently mutated in a variety of tumors, including adult lower-grade gliomas, pediatric GBM pediatric adrenocortical carcinoma, osteosarcoma, and neuroblastoma [100]. Cancer cells with a loss of ATRX gene display large and bright telomeric DNA foci that are significantly correlated with ALT [85], suggesting that ATRX may be a suppressor of the ALT mechanism and a good prognostic factor in cancers, such as in GBMs [101]. Moreover, the forced expression of ATRX in ALT-positive and ATRX-negative cell lines abolishes ALT-associated phenotypes [102][103], but ATRX, either by knocking out or knocking down in telomerase-positive cell lines did not present similar findings [96][103][104][105][106], suggesting that an ATRX loss alone is not sufficient for ALT activation. DAXX was originally identified as a Fas death receptor binding protein that induced apoptosis via JNK pathway activation. Thus, it was coined the death-domain-associated protein, DAXX [107]. ATRX and DAXX were initially seemed to be irrelevant. Analyzing H3.3 chaperone complexes identified ATRX and DAXX [108][109][110]. ATRX, in collaboration with DAXX, deposits H3.3 into telomeric and pericentromeric chromatin to prevent the formation of G-quadruplex DNA (G-4 DNA), a type of DNA structure that promotes homologous recombination repair and DNA-repair mechanisms, leading to telomere shortening [85][102][111]. These results reinforce the value of the ATRX/DAXX/H3.3 complex in ALT suppression (Figure 2). Gliomas with a WT TERT promoter frequently harbor mutations of ATRX to activate ALT [76]. Yang et al. successfully switched the telomere lengthening machinery of telomerase-positive cancer cells (HTC75) to an ALT-mediated telomere elongation mechanism by knocking out the TERT, inducing telomeric DNA damage, and disrupting the ATRX/DAXX complex [112], suggesting that ATRX and TERT mutations are mutually exclusive in conferring a selective growth advantage in cancers through telomere maintenance. Therefore, effective treatments should not only target TA in cancer cells but should also be aimed atmodulatingthe proper function of the ATRX/DAXX/H3.3 complex to destroy tumor cells.
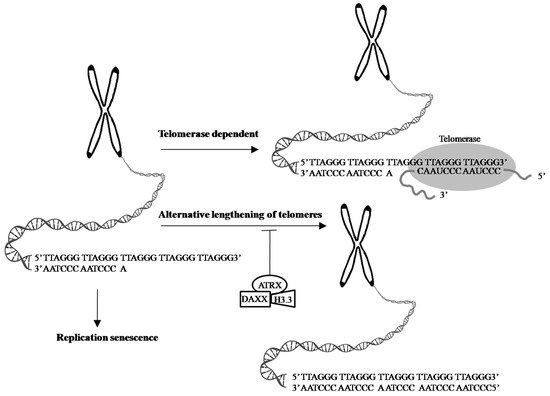
Figure 2. Telomeres are cap-like features at the ends of chromosomes that help protect them when cells divide. Telomeres contain thousands of repeats of the six-nucleotide sequence TTAGGG at the chromosome ends, with complementary DNA strand sequences AATCCC. 1. In the process of cell division, chromosome replication causes progressive telomere shortening, resulting in cell cycle arrest and apoptosis, leading to cellular senescence. 2. The shortening of telomeres can be counteracted by telomerase, which is made up of TERT, TERC, and specialized proteins. Telomerase maintains the length of telomeres stability by adding TTAGGG repeats to the end of the given chromosome, using its complementary TERC sequence as the template, together with TERT subunit as the catalytic component. 3. ALT is a phenomenon that 5–10% cancer cases exploit a telomerase-independent mechanism to elongate their telomeres [92]. The transformation from telomerase-dependent to ALT-mediated telomere lengthening may be one of strategies cancer cells adopt to escape cell senescence and apoptosis caused by telomerase dysfunction or absence [96][97]. ATRX, in collaboration with DAXX and H3.3 promotes the processes of telomere shortening [85][102][111]. Evidence shows the ATRX/DAXX/H3.3 complex in ALT suppression and a good prognostic factor in GBMs [101]. Arrow: possible pathway. T-bar: inhibition.
References
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812.
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773.
- Xu, X.; Zhao, J.; Xu, Z.; Peng, B.; Huang, Q.; Arnold, E.; Ding, J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 2004, 279, 33946–33957.
- Joseph, J.W.; Jensen, M.V.; Ilkayeva, O.; Palmieri, F.; Alarcon, C.; Rhodes, C.J.; Newgard, C.B. The mitochondrial citrate/isocitrate carrier plays a regulatory role in glucose-stimulated insulin secretion. J. Biol. Chem. 2006, 281, 35624–35632.
- Lee, S.H.; Jo, S.H.; Lee, S.M.; Koh, H.J.; Song, H.; Park, J.W.; Lee, W.H.; Huh, T.L. Role of NADP+-dependent isocitrate dehydrogenase (NADP+-ICDH) on cellular defence against oxidative injury by gamma-rays. Int. J. Radiat. Biol. 2004, 80, 635–642.
- Ward, P.S.; Patel, J.; Wise, D.R.; Abdel-Wahab, O.; Bennett, B.D.; Coller, H.A.; Cross, J.R.; Fantin, V.R.; Hedvat, C.V.; Perl, A.E.; et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010, 17, 225–234.
- Figueroa, M.E.; Abdel-Wahab, O.; Lu, C.; Ward, P.S.; Patel, J.; Shih, A.; Li, Y.; Bhagwat, N.; Vasanthakumar, A.; Fernandez, H.F.; et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010, 18, 553–567.
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744.
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.T.; Konermann, C.; Pfaff, E.; Tonjes, M.; Sill, M.; Bender, S.; et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012, 22, 425–437.
- Cairncross, J.G.; Ueki, K.; Zlatescu, M.C.; Lisle, D.K.; Finkelstein, D.M.; Hammond, R.R.; Silver, J.S.; Stark, P.C.; Macdonald, D.R.; Ino, Y.; et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl. Cancer Inst. 1998, 90, 1473–1479.
- Kraus, J.A.; Koopmann, J.; Kaskel, P.; Maintz, D.; Brandner, S.; Schramm, J.; Louis, D.N.; Wiestler, O.D.; von Deimling, A. Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J. Neuropathol. Exp. Neurol. 1995, 54, 91–95.
- Polivka, J., Jr.; Polivka, J.; Repik, T.; Rohan, V.; Hes, O.; Topolcan, O. Co-deletion of 1p/19q as Prognostic and Predictive Biomarker for Patients in West Bohemia with Anaplastic Oligodendroglioma. Anticancer Res. 2016, 36, 471–476.
- Reifenberger, J.; Reifenberger, G.; Liu, L.; James, C.D.; Wechsler, W.; Collins, V.P. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am. J. Pathol. 1994, 145, 1175–1190.
- Bettegowda, C.; Agrawal, N.; Jiao, Y.; Sausen, M.; Wood, L.D.; Hruban, R.H.; Rodriguez, F.J.; Cahill, D.P.; McLendon, R.; Riggins, G.; et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011, 333, 1453–1455.
- Jiao, Y.; Killela, P.J.; Reitman, Z.J.; Rasheed, A.B.; Heaphy, C.M.; de Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Oba-Shinjo, S.M.; Nagahashi Marie, S.K.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012, 3, 709–722.
- Labussiere, M.; Idbaih, A.; Wang, X.W.; Marie, Y.; Boisselier, B.; Falet, C.; Paris, S.; Laffaire, J.; Carpentier, C.; Criniere, E.; et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology 2010, 74, 1886–1890.
- Duan, J.; Bao, X.; Ma, X.; Zhang, Y.; Ni, D.; Wang, H.; Zhang, F.; Du, Q.; Fan, Y.; Chen, J.; et al. Upregulation of Far Upstream Element-Binding Protein 1 (FUBP1) Promotes Tumor Proliferation and Tumorigenesis of Clear Cell Renal Cell Carcinoma. PLoS ONE 2017, 12, e0169852.
- Han, F.; Zhang, J.; Ma, S.; Chen, X.; Liu, W.; He, X.; Fei, Z.; Wang, Y. Altered capicua transcriptional repressor gene expression exhibits distinct prognostic value for isocitrate dehydrogenase-mutant oligodendroglial tumors. Oncol. Lett. 2018, 15, 1459–1468.
- Cairncross, G.; Wang, M.; Shaw, E.; Jenkins, R.; Brachman, D.; Buckner, J.; Fink, K.; Souhami, L.; Laperriere, N.; Curran, W.; et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: Long-term results of RTOG 9402. J. Clin. Oncol. 2013, 31, 337–343.
- van den Bent, M.J.; Carpentier, A.F.; Brandes, A.A.; Sanson, M.; Taphoorn, M.J.; Bernsen, H.J.; Frenay, M.; Tijssen, C.C.; Grisold, W.; Sipos, L.; et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: A randomized European Organisation for Research and Treatment of Cancer phase III trial. J. Clin. Oncol. 2006, 24, 2715–2722.
- Kaneshiro, D.; Kobayashi, T.; Chao, S.T.; Suh, J.; Prayson, R.A. Chromosome 1p and 19q deletions in glioblastoma multiforme. Appl. Immunohistochem. Mol. Morphol. 2009, 17, 512–516.
- Nakamura, M.; Yang, F.; Fujisawa, H.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Loss of heterozygosity on chromosome 19 in secondary glioblastomas. J. Neuropathol. Exp. Neurol. 2000, 59, 539–543.
- Ohgaki, H.; Kleihues, P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007, 170, 1445–1453.
- Filipescu, D.; Muller, S.; Almouzni, G. Histone H3 variants and their chaperones during development and disease: Contributing to epigenetic control. Annu. Rev. Cell Dev. Biol. 2014, 30, 615–646.
- Saleem, M.; Abbas, K.; Manan, M.; Ijaz, H.; Ahmed, B.; Ali, M.; Hanif, M.; Farooqi, A.A.; Qadir, M.I. Review-Epigenetic therapy for cancer. Pak. J. Pharm. Sci. 2015, 28, 1023–1032.
- Szenker, E.; Ray-Gallet, D.; Almouzni, G. The double face of the histone variant H3.3. Cell Res. 2011, 21, 421–434.
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395.
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012, 44, 251–253.
- Bender, S.; Tang, Y.; Lindroth, A.M.; Hovestadt, V.; Jones, D.T.; Kool, M.; Zapatka, M.; Northcott, P.A.; Sturm, D.; Wang, W.; et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 2013, 24, 660–672.
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820.
- Buczkowicz, P.; Hoeman, C.; Rakopoulos, P.; Pajovic, S.; Letourneau, L.; Dzamba, M.; Morrison, A.; Lewis, P.; Bouffet, E.; Bartels, U.; et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat. Genet. 2014, 46, 451–456.
- Fang, J.; Huang, Y.; Mao, G.; Yang, S.; Rennert, G.; Gu, L.; Li, H.; Li, G.M. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSalpha interaction. Proc. Natl. Acad. Sci. USA 2018, 115, 9598–9603.
- Bjerke, L.; Mackay, A.; Nandhabalan, M.; Burford, A.; Jury, A.; Popov, S.; Bax, D.A.; Carvalho, D.; Taylor, K.R.; Vinci, M.; et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013, 3, 512–519.
- Martin, C.L.L. DH Telomeres. In Handbook of Human Molecular Evolution; John Wiley & Sons: Hoboken, NJ, USA, 2008; pp. 721–724.
- Ruiz-Herrera, A.; Nergadze, S.G.; Santagostino, M.; Giulotto, E. Telomeric repeats far from the ends: Mechanisms of origin and role in evolution. Cytogenet. Genome Res. 2008, 122, 219–228.
- Liu, D.; O’Connor, M.S.; Qin, J.; Songyang, Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004, 279, 51338–51342.
- Blasco, M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007, 3, 640–649.
- McClintock, B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics 1941, 26, 234–282.
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181.
- Gong, J.G.; Costanzo, A.; Yang, H.Q.; Melino, G.; Kaelin, W.G., Jr.; Levrero, M.; Wang, J.Y. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 1999, 399, 806–809.
- Stiewe, T.; Putzer, B.M. p73 in apoptosis. Apoptosis 2001, 6, 447–452.
- Vaziri, H.; Dragowska, W.; Allsopp, R.C.; Thomas, T.E.; Harley, C.B.; Lansdorp, P.M. Evidence for a mitotic clock in human hematopoietic stem cells: Loss of telomeric DNA with age. Proc. Natl. Acad. Sci. USA 1994, 91, 9857–9860.
- Brouilette, S.; Singh, R.K.; Thompson, J.R.; Goodall, A.H.; Samani, N.J. White cell telomere length and risk of premature myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 842–846.
- Valdes, A.M.; Andrew, T.; Gardner, J.P.; Kimura, M.; Oelsner, E.; Cherkas, L.F.; Aviv, A.; Spector, T.D. Obesity, cigarette smoking, and telomere length in women. Lancet 2005, 366, 662–664.
- Frenck, R.W., Jr.; Blackburn, E.H.; Shannon, K.M. The rate of telomere sequence loss in human leukocytes varies with age. Proc. Natl. Acad. Sci. USA 1998, 95, 5607–5610.
- Dalgard, C.; Benetos, A.; Verhulst, S.; Labat, C.; Kark, J.D.; Christensen, K.; Kimura, M.; Kyvik, K.O.; Aviv, A. Leukocyte telomere length dynamics in women and men: Menopause vs age effects. Int. J. Epidemiol. 2015, 44, 1688–1695.
- Benetti, R.; Garcia-Cao, M.; Blasco, M.A. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007, 39, 243–250.
- Celli, G.B.; de Lange, T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005, 7, 712–718.
- Steinert, S.; Shay, J.W.; Wright, W.E. Modification of subtelomeric DNA. Mol. Cell. Biol. 2004, 24, 4571–4580.
- Adams, J.; Martin-Ruiz, C.; Pearce, M.S.; White, M.; Parker, L.; von Zglinicki, T. No association between socio-economic status and white blood cell telomere length. Aging Cell 2007, 6, 125–128.
- Cherkas, L.F.; Hunkin, J.L.; Kato, B.S.; Richards, J.B.; Gardner, J.P.; Surdulescu, G.L.; Kimura, M.; Lu, X.; Spector, T.D.; Aviv, A. The association between physical activity in leisure time and leukocyte telomere length. Arch. Intern. Med. 2008, 168, 154–158.
- Nordfjall, K.; Eliasson, M.; Stegmayr, B.; Melander, O.; Nilsson, P.; Roos, G. Telomere length is associated with obesity parameters but with a gender difference. Obesity 2008, 16, 2682–2689.
- van der Harst, P.; van der Steege, G.; de Boer, R.A.; Voors, A.A.; Hall, A.S.; Mulder, M.J.; van Gilst, W.H.; van Veldhuisen, D.J.; MERIT-HF Study Group. Telomere length of circulating leukocytes is decreased in patients with chronic heart failure. J. Am. Coll. Cardiol. 2007, 49, 1459–1464.
- Brouilette, S.W.; Moore, J.S.; McMahon, A.D.; Thompson, J.R.; Ford, I.; Shepherd, J.; Packard, C.J.; Samani, N.J.; West of Scotland Coronary Prevention Study Group. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: A nested case-control study. Lancet 2007, 369, 107–114.
- Fitzpatrick, A.L.; Kronmal, R.A.; Gardner, J.P.; Psaty, B.M.; Jenny, N.S.; Tracy, R.P.; Walston, J.; Kimura, M.; Aviv, A. Leukocyte telomere length and cardiovascular disease in the cardiovascular health study. Am. J. Epidemiol. 2007, 165, 14–21.
- Zee, R.Y.; Michaud, S.E.; Germer, S.; Ridker, P.M. Association of shorter mean telomere length with risk of incident myocardial infarction: A prospective, nested case-control approach. Clin. Chim. Acta 2009, 403, 139–141.
- Sampson, M.J.; Winterbone, M.S.; Hughes, J.C.; Dozio, N.; Hughes, D.A. Monocyte telomere shortening and oxidative DNA damage in type 2 diabetes. Diabetes Care 2006, 29, 283–289.
- Valdes, A.M.; Richards, J.B.; Gardner, J.P.; Swaminathan, R.; Kimura, M.; Xiaobin, L.; Aviv, A.; Spector, T.D. Telomere length in leukocytes correlates with bone mineral density and is shorter in women with osteoporosis. Osteoporos. Int. 2007, 18, 1203–1210.
- Cawthon, R.M.; Smith, K.R.; O’Brien, E.; Sivatchenko, A.; Kerber, R.A. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 2003, 361, 393–395.
- Farzaneh-Far, R.; Cawthon, R.M.; Na, B.; Browner, W.S.; Schiller, N.B.; Whooley, M.A. Prognostic value of leukocyte telomere length in patients with stable coronary artery disease: Data from the Heart and Soul Study. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1379–1384.
- Yang, Z.; Huang, X.; Jiang, H.; Zhang, Y.; Liu, H.; Qin, C.; Eisner, G.M.; Jose, P.A.; Rudolph, L.; Ju, Z. Short telomeres and prognosis of hypertension in a chinese population. Hypertension 2009, 53, 639–645.
- Chin, L.; Artandi, S.E.; Shen, Q.; Tam, A.; Lee, S.L.; Gottlieb, G.J.; Greider, C.W.; DePinho, R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 1999, 97, 527–538.
- De Lange, T. Telomere-related genome instability in cancer. Cold Spring Harb. Symp. Quant. Biol. 2005, 70, 197–204.
- Meeker, A.K. Telomeres and telomerase in prostatic intraepithelial neoplasia and prostate cancer biology. Urol. Oncol. 2006, 24, 122–130.
- Shay, J.W.; Wright, W.E. Role of telomeres and telomerase in cancer. Semin. Cancer Biol. 2011, 21, 349–353.
- Wu, K.; Higashi, N.; Hansen, E.R.; Lund, M.; Bang, K.; Thestrup-Pedersen, K. Telomerase activity is increased and telomere length shortened in T cells from blood of patients with atopic dermatitis and psoriasis. J. Immunol. 2000, 165, 4742–4747.
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413.
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626.
- Shippen-Lentz, D.; Blackburn, E.H. Functional evidence for an RNA template in telomerase. Science 1990, 247, 546–552.
- Hiyama, K.; Hirai, Y.; Kyoizumi, S.; Akiyama, M.; Hiyama, E.; Piatyszek, M.A.; Shay, J.W.; Ishioka, S.; Yamakido, M. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J. Immunol. 1995, 155, 3711–3715.
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015.
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053.
- Feng, J.; Funk, W.D.; Wang, S.S.; Weinrich, S.L.; Avilion, A.A.; Chiu, C.P.; Adams, R.R.; Chang, E.; Allsopp, R.C.; Yu, J.; et al. The RNA component of human telomerase. Science 1995, 269, 1236–1241.
- Mocellin, S.; Pooley, K.A.; Nitti, D. Telomerase and the search for the end of cancer. Trends Mol. Med. 2013, 19, 125–133.
- Smekalova, E.M.; Shubernetskaya, O.S.; Zvereva, M.I.; Gromenko, E.V.; Rubtsova, M.P.; Dontsova, O.A. Telomerase RNA biosynthesis and processing. Biochemistry 2012, 77, 1120–1128.
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A.; Jr Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026.
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961.
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959.
- Liu, X.; Wu, G.; Shan, Y.; Hartmann, C.; von Deimling, A.; Xing, M. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle 2013, 12, 1637–1638.
- Arita, H.; Narita, Y.; Takami, H.; Fukushima, S.; Matsushita, Y.; Yoshida, A.; Miyakita, Y.; Ohno, M.; Shibui, S.; Ichimura, K. TERT promoter mutations rather than methylation are the main mechanism for TERT upregulation in adult gliomas. Acta Neuropathol. 2013, 126, 939–941.
- Koelsche, C.; Sahm, F.; Capper, D.; Reuss, D.; Sturm, D.; Jones, D.T.; Kool, M.; Northcott, P.A.; Wiestler, B.; Bohmer, K.; et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013, 126, 907–915.
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477.
- Nonoguchi, N.; Ohta, T.; Oh, J.E.; Kim, Y.H.; Kleihues, P.; Ohgaki, H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013, 126, 931–937.
- Vinagre, J.; Almeida, A.; Populo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185.
- Heaphy, C.M.; de Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011, 333, 425.
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.; Tonjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231.
- Molano, M.; Martin, D.C.; Moreno-Acosta, P.; Hernandez, G.; Cornall, A.; Buitrago, O.; Gamboa, O.; Garland, S.; Tabrizi, S.; Munoz, N. Telomerase activity in cervical scrapes of women with high-grade cervical disease: A nested case-control study. Oncol. Lett. 2018, 15, 354–360.
- Wang, N.; Xu, D.; Sofiadis, A.; Hoog, A.; Vukojevic, V.; Backdahl, M.; Zedenius, J.; Larsson, C. Telomerase-dependent and independent telomere maintenance and its clinical implications in medullary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2014, 99, E1571–E1579.
- Bornstein-Quevedo, L.; Garcia-Hernandez, M.L.; Camacho-Arroyo, I.; Herrera, M.F.; Angeles, A.A.; Trevino, O.G.; Gamboa-Dominguez, A. Telomerase activity in well-differentiated papillary thyroid carcinoma correlates with advanced clinical stage of the disease. Endocr. Pathol. 2003, 14, 213–219.
- Fernandez-Marcelo, T.; Gomez, A.; Pascua, I.; de Juan, C.; Head, J.; Hernando, F.; Jarabo, J.R.; Calatayud, J.; Torres-Garcia, A.J.; Iniesta, P. Telomere length and telomerase activity in non-small cell lung cancer prognosis: Clinical usefulness of a specific telomere status. J. Exp. Clin. Cancer Res. 2015, 34, 78.
- Satyanarayana, A.; Manns, M.P.; Rudolph, K.L. Telomeres and telomerase: A dual role in hepatocarcinogenesis. Hepatology 2004, 40, 276–283.
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997, 3, 1271–1274.
- Slatter, T.L.; Tan, X.; Yuen, Y.C.; Gunningham, S.; Ma, S.S.; Daly, E.; Packer, S.; Devenish, C.; Royds, J.A.; Hung, N.A. The alternative lengthening of telomeres pathway may operate in non-neoplastic human cells. J. Pathol. 2012, 226, 509–518.
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011, 179, 1608–1615.
- Wiestler, B.; Capper, D.; Holland-Letz, T.; Korshunov, A.; von Deimling, A.; Pfister, S.M.; Platten, M.; Weller, M.; Wick, W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013, 126, 443–451.
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suva, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273–277.
- Shay, J.W.; Reddel, R.R.; Wright, W.E. Cancer. Cancer and telomeres—An ALTernative to telomerase. Science 2012, 336, 1388–1390.
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330.
- Gibbons, R. Alpha thalassaemia-mental retardation, X linked. Orphanet J. Rare Dis. 2006, 1, 15.
- Dyer, M.A.; Qadeer, Z.A.; Valle-Garcia, D.; Bernstein, E. ATRX and DAXX: Mechanisms and Mutations. Cold Spring Harb. Perspect. Med. 2017, 7, a026567.
- Pekmezci, M.; Rice, T.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Hansen, H.; Sicotte, H.; Kollmeyer, T.M.; McCoy, L.S.; Sarkar, G.; et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: Additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017, 133, 1001–1016.
- Clynes, D.; Higgs, D.R.; Gibbons, R.J. The chromatin remodeller ATRX: A repeat offender in human disease. Trends Biochem. Sci. 2013, 38, 461–466.
- Napier, C.E.; Huschtscha, L.I.; Harvey, A.; Bower, K.; Noble, J.R.; Hendrickson, E.A.; Reddel, R.R. ATRX represses alternative lengthening of telomeres. Oncotarget 2015, 6, 16543–16558.
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Taylor, S.; Mitson, M.; Bachrati, C.Z.; Higgs, D.R.; Gibbons, R.J. ATRX dysfunction induces replication defects in primary mouse cells. PLoS ONE 2014, 9, e92915.
- Eid, R.; Demattei, M.V.; Episkopou, H.; Auge-Gouillou, C.; Decottignies, A.; Grandin, N.; Charbonneau, M. Genetic Inactivation of ATRX Leads to a Decrease in the Amount of Telomeric Cohesin and Level of Telomere Transcription in Human Glioma Cells. Mol. Cell. Biol. 2015, 35, 2818–2830.
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012, 8, e1002772.
- Yang, X.; Khosravi-Far, R.; Chang, H.Y.; Baltimore, D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 1997, 89, 1067–1076.
- Drane, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010, 24, 1253–1265.
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691.
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.M.; Stadler, S.C.; Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 2010, 107, 14075–14080.
- Amorim, J.P.; Santos, G.; Vinagre, J.; Soares, P. The Role of ATRX in the Alternative Lengthening of Telomeres (ALT) Phenotype. Genes 2016, 7, 66.
- Hu, Y.; Shi, G.; Zhang, L.; Li, F.; Jiang, Y.; Jiang, S.; Ma, W.; Zhao, Y.; Songyang, Z.; Huang, J. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci. Rep. 2016, 6, 32280.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
906
Revisions:
2 times
(View History)
Update Date:
03 Mar 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




