
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xiaohu Zhou | + 1845 word(s) | 1845 | 2022-01-10 02:32:56 | | | |
| 2 | Nora Tang | Meta information modification | 1845 | 2022-02-11 09:12:31 | | |
Video Upload Options
Single-cell analysis has become a powerful and indispensable tool in modern biological and medical research. Single-cell isolation is the key step for single-cell analysis. Single-cell printing could utilize various microfluidic technologies for single-cell isolation and analysis, such as droplet microfluidics, microwell arrays, and hydrodynamic traps. Single-cell printing shows several distinct advantages among the single-cell isolation techniques, such as precise deposition, high encapsulation efficiency, and easy recovery. With the development of single-cell printing in the past decade, various single-cell printing-based single-cell analyses and applications have been performed, ranging from single-cell array-based screening and single-cell-based mass spectroscopy to live three-dimensional tissue formation.
1. Introduction
As the key step for single-cell analysis, single-cell isolation has attracted great attention, and a number of techniques have been developed for single-cell isolation and manipulation [1]. Single-cell printing could utilize various microfluidic technologies for single-cell isolation and analysis, such as droplet microfluidics [2][3][4], microwell arrays [5][6], and hydrodynamic traps [7][8]. Single-cell printing has several distinct advantages. First, single-cell printing can effectively and precisely deposit cells at specific sites in high throughput [9]. Second, the printed single cells or colonies can be easily recovered with addressability for subsequent analysis. Third, it is convenient to integrate the highly efficient single-cell printing with other techniques, such as imaging system [10][11], electric field [12][13][14], and acoustic field [15][16], and the single-cell encapsulation efficiency can reach more than 90%. Furthermore, bioprinting with the single-cell resolution can not only print 2D single-cell arrays for single-cell analysis but also three-dimensional tissue matrixes and organs for tissue engineering, drug discovery, and toxicology [17][18]. With the development of single-cell printing in the past decade, various single-cell printing-based single-cell analyses and applications have been performed, ranging from single-cell array-based screening [19][20][21][22][23][24] and single-cell-based mass spectroscopy [25][26][27][28][29][30] to live three-dimensional tissue formation [17][18][31].
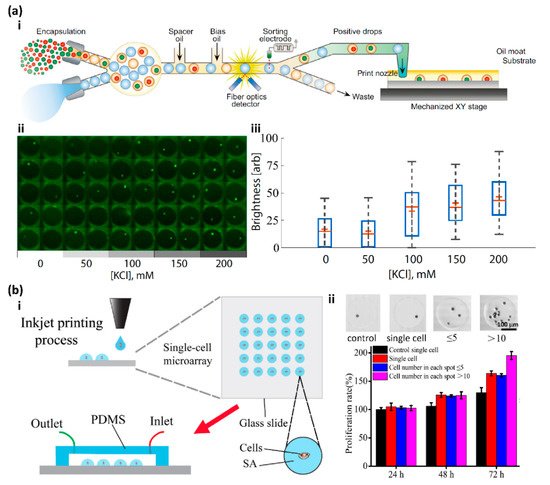
2. High Throughput Screening
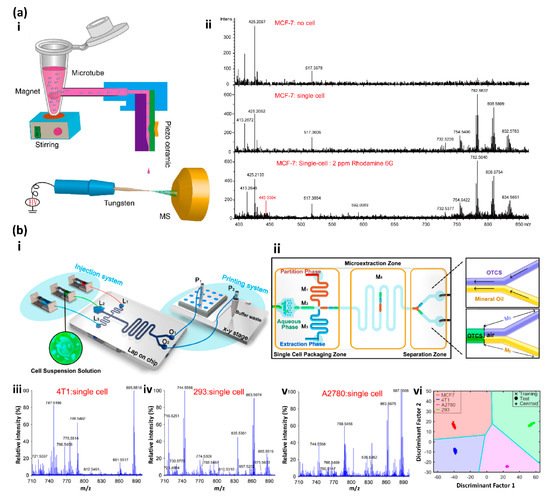
3. Mass Spectrometry-Based Single-Cell Analysis
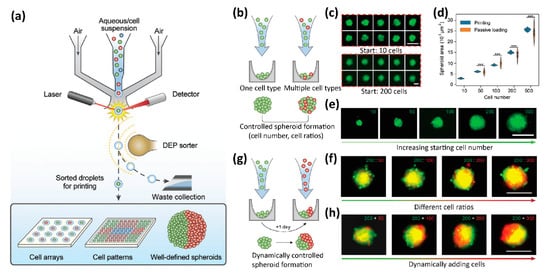
4. 3D Tissue Printing
5. Summary and Future Perspective
Cell bioprinting has made remarkable progress in printing three-dimensional multicellular tissues and organs in the past decade [17][18][39]. However, compared to the 3D bioprinting, where the single-cell resolution is not necessarily required, there is little progress in single-cell printing-based single-cell analysis, although the recently developed single-cell bioprinting strategies show the great potential of single-cell analysis in-depth with promising advantages, such as high encapsulation efficiency, precise deposition, and easy recovery. Currently, the most common strategies for single-cell analysis (omics) are based on droplet microfluidics [3]. Several issues limit the applications of current single-cell printing for in-depth single-cell analysis. First and foremost, although the reliability and efficiency of the encapsulation of single cells are dramatically enhanced, the overall throughput of the single-cell printing is still low, at a rate of ~2 Hz [42]. Second, compared to the popular microfluidic technologies in an isolated system, current printing strategies normally rely on printing the single-cell droplets in an open environment, which may cause interference and deviation in the subsequent analysis. Third, many subsequent analyses after single-cell isolation are performed on instruments that are incompatible with single-cell printing [43][44].
Despite the above issues, single-cell printing is experiencing rapid development. Several commercial single-cell printers are now available. Throughput, single-cell efficiency, robustness, and reproducibility are among the most important factors to consider when evaluating a single-cell printing technique or product. With the increasing demand in tissue engineering, precision medicine, liquid biopsy, and drug discovery, we expect that single-cell printing will play a more vital role in single-cell analysis in the future.
References
- Zhang, X.; Wei, X.; Wei, Y.; Chen, M.; Wang, J. The Up-to-Date Strategies for the Isolation and Manipulation of Single Cells. Talanta 2020, 218, 121147.
- Zhang, X.; Li, T.; Liu, F.; Chen, Y.; Yao, J.; Li, Z.; Huang, Y.; Wang, J. Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems. Mol. Cell 2019, 73, 130–142.
- Matuła, K.; Rivello, F.; Huck, W.T.S. Single-Cell Analysis Using Droplet Microfluidics. Adv. Biosyst. 2020, 4, 1900188.
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell 2015, 161, 1187–1201.
- Han, X.; Wang, R.; Zhou, Y.; Fei, L.; Sun, H.; Lai, S.; Saadatpour, A.; Zhou, Z.; Chen, H.; Ye, F.; et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 2018, 172, 1091–1107.
- Rettig, J.R.; Folch, A. Large-Scale Single-Cell Trapping And Imaging Using Microwell Arrays. Anal. Chem. 2005, 77, 5628–5634.
- Zhou, Y.; Basu, S.; Wohlfahrt, K.J.; Lee, S.F.; Klenerman, D.; Laue, E.D.; Seshia, A.A. A Microfluidic Platform for Trapping, Releasing and Super-Resolution Imaging of Single Cells. Sens. Actuators B Chem. 2016, 232, 680–691.
- Tan, W.H.; Takeuchi, S. A Trap-and-Release Integrated Microfluidic System for Dynamic Microarray Applications. Proc. Natl. Acad. Sci. USA 2007, 104, 1146–1151.
- Calvert, P. Printing Cells. Science 2007, 318, 208–209.
- Yusof, A.; Keegan, H.; Spillane, C.D.; Sheils, O.M.; Martin, C.M.; O’Leary, J.J.; Zengerle, R.; Koltay, P. Inkjet-like Printing of Single-Cells. Lab Chip 2011, 11, 2447–2454.
- Wang, Y.; Wang, X.; Pan, T.; Li, B.; Chu, J. Label-Free Single-Cell Isolation Enabled by Microfluidic Impact Printing and Real-Time Cellular Recognition. Lab Chip 2021, 21, 3695–3706.
- Schoendube, J.; Wright, D.; Zengerle, R.; Koltay, P. Single-Cell Printing Based on Impedance Detection. Biomicrofluidics 2015, 9, 014117.
- Feng, L.; Sun, Y.; Ohsumi, C.; Arai, F. Accurate Dispensing System for Single Oocytes Using Air Ejection. Biomicrofluidics 2013, 7, 054113.
- Nagai, M.; Kato, K.; Soga, S.; Santra, T.S.; Shibata, T. Scalable Parallel Manipulation of Single Cells Using Micronozzle Array Integrated with Bidirectional Electrokinetic Pumps. Micromachines 2020, 11, 442.
- Demirci, U.; Montesano, G. Single Cell Epitaxy by Acoustic Picolitre Droplets. Lab Chip 2007, 7, 1139–1145.
- Guo, F.; Mao, Z.; Chen, Y.; Xie, Z.; Lata, J.P.; Li, P.; Ren, L.; Liu, J.; Yang, J.; Dao, M.; et al. Three-Dimensional Manipulation of Single Cells Using Surface Acoustic Waves. Proc. Natl. Acad. Sci. USA 2016, 113, 1522–1527.
- Bertassoni, L.E. Bioprinting of Complex Multicellular Organs with Advanced Functionality—Recent Progress and Challenges Ahead. Adv. Mater. 2021, 2101321.
- Daly, A.C.; Prendergast, M.E.; Hughes, A.J.; Burdick, J.A. Bioprinting for the Biologist. Cell 2021, 184, 18–32.
- Lin, X.; Fang, F.; Wang, C.; Kankala, R.K.; Zhou, S. Inkjet Printing-Assisted Single-Cell Microarray on a Hydrophobic Surface Chip for Real-Time Monitoring of Enzyme Kinetics at Single-Cell Level. Talanta 2021, 225, 122019.
- Sun, Y.; Song, W.; Sun, X.; Zhang, S. Inkjet-Printing Patterned Chip on Sticky Superhydrophobic Surface for High-Efficiency Single-Cell Array Trapping and Real-Time Observation of Cellular Apoptosis. ACS Appl. Mater. Interfaces 2018, 10, 31054–31060.
- Cole, R.H.; Tang, S.-Y.; Siltanen, C.A.; Shahi, P.; Zhang, J.Q.; Poust, S.; Gartner, Z.J.; Abate, A.R. Printed Droplet Microfluidics for on Demand Dispensing of Picoliter Droplets and Cells. Proc. Natl. Acad. Sci. USA 2017, 114, 8728–8733.
- Zhang, K.; Chou, C.-K.; Xia, X.; Hung, M.-C.; Qin, L. Block-Cell-Printing for Live Single-Cell Printing. Proc. Natl. Acad. Sci. USA 2014, 111, 2948–2953.
- Stumpf, F.; Schoendube, J.; Gross, A.; Rath, C.; Niekrawietz, S.; Koltay, P.; Roth, G. Single-Cell PCR of Genomic DNA Enabled by Automated Single-Cell Printing for Cell Isolation. Biosens. Bioelectron. 2015, 69, 301–306.
- Yim, M.; Shaw, D. Achieving Greater Efficiency and Higher Confidence in Single-Cell Cloning by Combining Cell Printing and Plate Imaging Technologies. Biotechnol. Prog. 2018, 34, 1454–1459.
- Li, Q.; Tang, F.; Huo, X.; Huang, X.; Zhang, Y.; Wang, X.; Zhang, X. Native State Single-Cell Printing System and Analysis for Matrix Effects. Anal. Chem. 2019, 91, 8115–8122.
- Ellis, S.R.; Ferris, C.J.; Gilmore, K.J.; Mitchell, T.W.; Blanksby, S.J.; Panhuis, M. In het Direct Lipid Profiling of Single Cells from Inkjet Printed Microarrays. Anal. Chem. 2012, 84, 9679–9683.
- Cahill, J.F.; Riba, J.; Kertesz, V. Rapid, Untargeted Chemical Profiling of Single Cells in Their Native Environment. Anal. Chem. 2019, 91, 6118–6126.
- Chen, F.; Lin, L.; Zhang, J.; He, Z.; Uchiyama, K.; Lin, J.M. Single-Cell Analysis Using Drop-on-Demand Inkjet Printing and Probe Electrospray Ionization Mass Spectrometry. Anal. Chem. 2016, 88, 4354–4360.
- Kazoe, Y.; Shimizu, Y.; Morikawa, K.; Terui, Y.; Irie, T.; Kitamori, T. Development of Microfluidic Droplet Shooter and Its Application to Interface for Mass Spectrometry. Sens. Actuators B Chem. 2021, 340, 129957.
- Luo, C.; Ma, Y.; Li, H.; Chen, F.; Uchiyama, K.; Lin, J.M. Generation of Picoliter Droplets of Liquid for Electrospray Ionization with Piezoelectric Inkjet. J. Mass Spectrom. 2013, 48, 321–328.
- Zhang, P.; Abate, A.R. High-Definition Single-Cell Printing: Cell-by-Cell Fabrication of Biological Structures. Adv. Mater. 2020, 32, 2005346.
- Yin, L.; Zhang, Z.; Liu, Y.; Gao, Y.; Gu, J. Recent Advances in Single-Cell Analysis by Mass Spectrometry. Analyst 2019, 144, 824–845.
- Perkel, J.M. Single-Cell Proteomics Takes Centre Stage. Nature 2021, 597, 580–582.
- Budnik, B.; Levy, E.; Harmange, G.; Slavov, N. SCoPE-MS: Mass Spectrometry of Single Mammalian Cells Quantifies Proteome Heterogeneity during Cell Differentiation. Genome Biol. 2018, 19, 1–12.
- Slavov, N. Single-Cell Protein Analysis by Mass Spectrometry. Curr. Opin. Chem. Biol. 2021, 60, 1–9.
- Greenberg, A.S.; Coleman, R.A.; Kraemer, F.B.; McManaman, J.L.; Obin, M.S.; Puri, V.; Yan, Q.W.; Miyoshi, H.; Mashek, D.G. The Role of Lipid Droplets in Metabolic Disease in Rodents and Humans. J. Clin. Investig. 2011, 121, 2102–2110.
- Kumar, P.; Ebbens, S.; Zhao, X. Inkjet Printing of Mammalian Cells—Theory and Applications. Bioprinting 2021, 23, e00157.
- Gao, G.; Kim, B.S.; Jang, J.; Cho, D.-W. Recent Strategies in Extrusion-Based Three-Dimensional Cell Printing toward Organ Biofabrication. ACS Biomater. Sci. Eng. 2019, 5, 1150–1169.
- Harley, W.S.; Li, C.C.; Toombs, J.; O’Connell, C.D.; Taylor, H.K.; Heath, D.E.; Collins, D.J. Advances in Biofabrication Techniques towards Functional Bioprinted Heterogeneous Engineered Tissues: A Comprehensive Review. Bioprinting 2021, 23, e00147.
- Murphy, S.V.; De Coppi, P.; Atala, A. Opportunities and Challenges of Translational 3D Bioprinting. Nat. Biomed. Eng. 2019, 4, 370–380.
- Murphy, S.V.; Atala, A. 3D Bioprinting of Tissues and Organs. Nat. Biotechnol. 2014, 32, 773–785.
- Chen, C.; Xu, D.; Bai, S.; Yu, Z.; Zhu, Y.; Xing, X.; Chen, H. Dynamic Screening and Printing of Single Cells Using a Microfluidic Chip with Dual Microvalves. Lab Chip 2020, 20, 1227–1237.
- Wen, L.; Tang, F. Boosting the Power of Single-Cell Analysis. Nat. Biotechnol. 2018, 36, 408–409.
- Stein, C.M.; Weiskirchen, R.; Damm, F.; Strzelecka, P.M. Single-Cell Omics: Overview, Analysis, and Application in Biomedical Science. J. Cell. Biochem. 2021, 122, 1571–1578.




