1. Introduction
The global challenge to prevent fungal spoilage and mycotoxin contamination on food and feed requires the development of new antifungal strategies. Given their multistep mode of action, the development of fungal resistance to AMPs is presumed to be slow or delayed compared to conventional fungicides. Interestingly, AMPs also accomplish important biological functions other than antifungal activity, including anti-mycotoxin biosynthesis activity, which opens novel aspects for their future use in agriculture and food industry to fight mycotoxin contamination. AMPs can reach intracellular targets and exert their activity by mechanisms other than membrane permeabilization. The mechanisms through which AMPs affect mycotoxin production are varied and complex, ranging from oxidative stress to specific inhibition of enzymatic components of mycotoxin biosynthetic pathways.
2. General Properties and Characteristics of Antimicrobial Peptides and Proteins (AMPs)
AMPs are small bioactive proteins or peptides, mostly cationic, that are naturally produced by nearly all living organisms. They primarily act as components of their innate immune system, becoming the first-line defense against microbial attacks in higher organisms. Additionally, AMPs might be produced as competition strategies by microorganisms to limit the growth of other competitors
[1][2]. AMPs are present in bacteria, fungi, plants, invertebrates and vertebrates
[3][4][5], and are known for their broad spectrum activity against bacteria, fungi, viruses, protozoa and/or even cancer cells
[6][7]. Remarkably, there are AMPs particularly effective against fungi
[3][8][9] and some of them show antifungal activity against mycotoxin-producing fungi.
AMPs are basically synthesized by two biosynthetic routes. Most are ribosomally encoded AMPs, while other AMPs are generated by non-ribosomal peptide synthases (NRPSs). The latter are mainly found in bacteria, in particular
Actinomycetes and
Bacilli [10]. The NRPS-generated AMPs are characterized by the incorporation of nonproteinogenic amino acids into the sequence and are often heavily modified through hydroxylation, glycosylation, lipidation, and cyclization
[11].
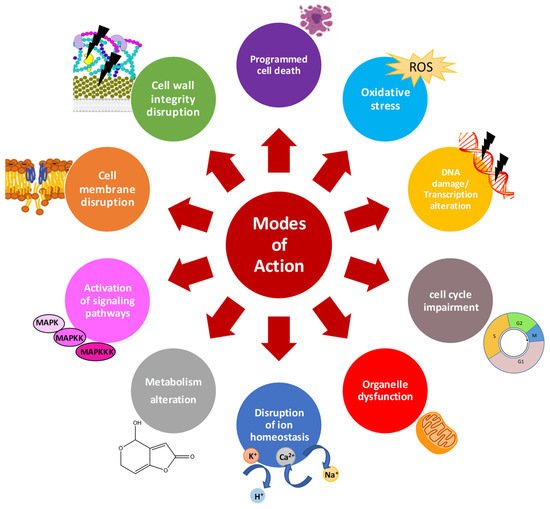
Characterization of the mode of action of AMPs is essential to improve their activity, avoid development of resistance, and accelerate their use as therapeutics or food preservatives. There is a significant volume of information available on the general mechanisms of action of AMPs (
Figure 1)
[12][13][14][15][16]. In general, AMPs can act at multiple cell targets. Cationic AMPs physically interact with the negatively charged microbial envelopes
[17]. However, electrostatic interactions cannot entirely explain other observed activity of AMPs, and thus, specific component of membrane envelopes seem to aid AMP interactions. In fungi, the cell wall plays a key role in the internalization and activity of several AMPs. Different AMPs has been reported to affect fungal cell wall by inhibiting β-glucan or chitin synthesis, and targeting mannoproteins from the cell wall in sensitive fungi
[8].
Figure 1. General mode of action of antifungal AMPs.
Once AMPs diffuse through the cell wall, they face the cell membrane. Any alteration of the plasma membrane may impact the distribution, regulation, activity and signaling function of membrane proteins, with adverse effects on fungal cells. Once the interaction occurs, AMPs are classified as membrane-disruptive or membrane non-disruptive. The cationic and amphipathic character of most AMPs allow the disruption of lipid cell membranes causing pore formation, loss of biophysical properties and cell killing
[12][18]. However, peptides acting through a lytic mechanisms are often highly toxic to different cell types
[15]. Therefore, AMPs with non-lytic mechanism such as the cell penetrating peptides (CPPs) are preferred
[13]. Once inside the cells, AMPs may target multiple processes. Common patterns of AMPs are the disruption of intracellular ion homeostasis, disruption of internal organelles such as mitochondria, (in)activation of signaling cascades, induction of reactive oxygen species (ROS) or apoptotic markers, disruption of cell cycle, DNA damage, and transcription and protein biosynthesis alteration
[19][15] (
Figure 1).
AMPs can also affect less commonly reported processes such as the production of secondary metabolites, including mycotoxins, which opens novel aspects for their future use in crops, postharvest and food processes. However, little is known about their mechanisms affecting mycotoxin biosynthesis in filamentous fungi. Studies on the effect on mycotoxin biosynthesis are restricted mainly to cyclopeptides derived from bacteria such as lipopeptides, small cysteine rich proteins (CRPs) which include defensins and antifungal AMPs of fungal origin (AFPs), and non-natural synthetic peptides. The main characteristics of these antifungal AMPs are summarized below.
Cyclopeptides derived from microorganisms contain both proteinogenic and unnatural amino acid residues
[20]. Among them, lipopeptides produced by members of the
Bacillus genus are compounds of great interest due to their activity against mycotoxin-producing fungi. These low molecular weight secondary metabolites have a broad range of activity, high biodegradability and low toxicity and are usually synthetized through NRPSs. They are composed of a hydrophilic cyclic peptide structure of 7–10 amino acids linked to a hydrophobic fatty acid chain with 13–19 carbon atoms. These compounds maintain their activities at high temperatures and different pH values; additionally, they resist peptidase and protease treatments
[21][22]. Lipopeptides are classified into three major families according to their amino acid sequence: iturins which are heptapeptides with a β-amino fatty acid; fengycins which are decapeptides with a β-hydroxy fatty acid chain and surfactins, heptapeptides containing a β-hydroxy fatty acid tail with synergistic action with the two previous groups.
Defensins found in mammals, insects and plants (45–54 amino acids in length) form by far, the largest family of CRPs and are highly active against a large range of microorganisms. Regardless of the origin, defensins are structurally similar peptides. They have β-hairpin structures, stabilized by three/four disulfide bonds, but their sequences are divergent and show different activities, which include antifungal, antibacterial, or antitumoral activities
[4]. Another CRP group of interest comprises of the AMPs of fungal origin, the so-called AFPs. AFPs are small (45–64 amino acids) and cationic defensin-like proteins that are produced and secreted to the culture medium by filamentous ascomycetes, mostly from the genera
Aspergillus and
Penicillium, and exhibit antifungal activity
[19]. Several of these proteins have activity towards fungal plant pathogens with minimal inhibitory concentration (MIC) in the low micromolar range
[23][24][25], and no toxicity to bacterial, plant or animal cells
[25][26][27]. AFPs fold into five β-strands forming two packed β-sheets that share a common interface, and typically have six cysteine residues, forming three disulfide bonds
[28][29]. A three-dimensional peptide signature, called the ɣ-core (Gly-X-Cys-X
3–9-Cys), is present in virtually all defensins and AFPs
[30].
Finally, the synthetic peptides with antifungal activities should also be noted. Synthetic AMPs are designed de novo based on the properties of natural AMPs or identified using combinatorial approaches. Peptide analogs of natural AMPs have been synthesized with substituted, deleted, or extended amino acids. Synthetic analogs have been produced through the modification of amino acid sequence, either by shortening the sequence to determine minimal antimicrobial motifs, or by extending peptide length, even by fusion of fragments from different peptides
[31]. These approaches, mainly directed to improve the antifungal activity, reduce toxicity to non-target cells and increase stability against degradation; additionally, they have contributed substantially to increasing the number and diversity of known AMPs
[3][32][33][34].
3. Effects of Distinct AMPs on Growth of Mycotoxin-Producing Fungi
Mycotoxins are secondary metabolites that are normally produced at the end of the exponential growth phase. Thus, mycotoxin production is generally thought to be correlated with the growth rate of producing fungi. Therefore, inhibiting fungal growth is often considered as the most effective strategy to prevent mycotoxin production.
Here we describe those antifungal AMPs that show activity against common mycotoxin-producing fungi, such as Alternaria, Aspergillus, Penicilllium and Fusarium species. These peptides have distinct and phylogenetically distant origins, ranging from microorganisms, to plants and mammals, as well as synthetic rationally designed peptides.
3.1. Antifungal AMPs from Microorganisms
A wide diversity of antifungal AMPs, produced by bacteria, are able to control fungal growth in vitro and in vivo. They mainly include antifungal AMPs produced by lactic acid bacteria (LAB), as well as species from the
Streptomyces,
Bacillus and
Burkholderia genera, which are particularly active against fungal species belonging to the
Aspergillus,
Penicillium and
Fusarium genera, but also to other species such as those from the
Byssochlamys genus (
Table 1). As examples of applications in vivo, the antifungal protein YvgO isolated from
Bacillus thuringiensis, was able to extend the shelf-life of different fruit juices inoculated with the PAT producer
Byssochlamys fulva, and provided a complimentary measure of protection in UV-treated fruit juices
[35]. On the other hand, a high antifungal activity of peptides generated by
L. plantarum TE10 was reported against
A. flavus. Results demonstrated promising application of the peptide mixture as bio-control agent to prevent the growth of
A. flavus in maize
[36]. Relevant bacterial AMPs with effect on mycotoxin biosynthesis are highlighted in the next section.
Fungi have a complex repertoire of AFPs that differ in amino acid composition and sequence
[3][37][27][38][39]. Several studies already indicated that some of the most hazardous mycotoxin-producing fungi are sensitive to AFPs (
Table 1). Of interest are those studies showing antifungal activity in a wide range of mycotoxin producers and differences in susceptibility to AFPs among the fungal genera and species. Delgado et al.
[40] evaluated the antifungal activity of PgAFP from
P. chrysogenum against toxigenic fungi commonly found in dry-ripened foods. PgAFP retarded the growth of most fungi tested and the main mycotoxin-producing fungi analyzed, such as those producing AFs (
A. flavus and
Aspergillus parasiticus), OTA (
A. carbonarius,
A. ochraceus, and
P. nordicum), ST (
A. versicolor) and PAT (
P. expansum and
P. griseofulvum). Recently, AFPs from
P. digitatum (PdAfpB) and
P. expansum (PeAfpA, PeAfpB and PeAfpC), were tested against a representative panel of mycotoxin-producing fungi belonging to the genera
Alternaria,
Aspergillus,
Byssochlamys,
Fusarium and
Penicillium [41]. These were previously reported to produce up to 26 different mycotoxins. AFPs showed significant activity against most of the mycotoxigenic fungi tested, in particular PeAfpA. PeAfpC showed powerful inhibition against
Byssochlamys spectabilis (PAT producer), which is an important spoilage fungus in pasteurized food products, such as fruit juices and canned fruits
[42]. Differences in susceptibility to AFPs were observed among fungal genera. In general,
Aspergillus,
Byssochlamys and
Penicillium were more sensitive than the
Fusarium genus. Moreover, the antifungal effect of AFPs also differed within the same species
[40][41][43]. Further studies on susceptibility and resistance of fungal species including more strains from each species are needed to elucidate antifungal specificities of AFPs.
Table 1. Microbial antifungal proteins and peptides with activity against mycotoxin-producing fungi.
| Origin |
Peptide |
Target Fungi |
Ref. |
| Bacteria |
|
|
|
| Bacillus amyloliquefaciens |
Flagellin |
F. oxysporum, A. niger |
[44] |
| B. subtilis |
Fengycins |
F. oxysporum |
[45] |
| B. subtilis |
Iturin A |
Aspergillus spp., Fusarium spp., Penicilium spp. |
[46] |
| B. thuringiensis |
YvgO |
B. fulva |
[35] |
| Burkholderia cepacia |
Cepacidines |
A. niger |
[47] |
| Enterococcus durans |
Duracin |
F. culmorum |
[48] |
| Lactic acid bacteria |
Bacteriocins |
A. parasiticus, P. expansum |
[49][50] |
| Lactobacillus brevis AM7 |
Peptides |
P. roqueforti |
[51] |
| L. paracasei |
Bacteriocin F1 |
P. glaucum, A. niger, A. flavus |
[52] |
| L. plantarum |
LR/14 |
A. niger, P. chrysogenum |
[53] |
| L. plantarum |
FPSHTGMSVPPP |
Aspergillus spp., P. roqueforti |
[54] |
| L. plantarum TE10 |
Peptides MIX |
A. flavus |
[36] |
| Streptomyces spp. |
C/33-6 |
F. graminearum |
[55] |
| S. tendae |
Nikkomycin Z |
Aspergillus spp., Fusarium spp., Penicilium spp. |
[56] |
| S. tendae Tu901 |
AFP1 |
A. fumigatus |
[57] |
| Fungi |
|
|
|
| Aspergillus giganteous |
AFP |
Fusarium spp. |
[58] |
| A. clavatus |
AcAFP |
F. oxysporum, F. solani |
[59] |
| A. clavatus |
AcAMP |
F. oxysporum, F. solani |
[60] |
| A. niger |
Anafp |
A. flavus, F. oxysporum, F. solani |
[23] |
| Fusarium graminearum |
FgAFP |
F. verticilloides, F. proliferatum |
[61] |
| Emericellopsis alkalina |
Emericellipsin A |
A.niger, A. flavus |
[62] |
| Monascus pilosus |
MAFP1 |
Fusarium spp. |
[63] |
| Neosartoria fischeri |
NFAP |
A. nidulans, F. graminearum |
[64] |
| N. fischeri |
NFAP2 |
A. nidulans |
[65] |
| Penicillium citrinum |
PcPAF |
F. oxysporum |
[66] |
| P. chrysogenum |
PAF |
F. oxysporum, A. flavus |
[67] |
| P. chrysogenum |
PgAFP/PAFB |
F. oxysporum, A. flavus |
[40][68] |
| P. chrysogenum |
Pc-Arctin/PAFC |
A. longipes, B. spectabilis |
[24][69] |
| P. digitatum |
PdAfpB |
F. oxysporum, P. expansum |
[41][70] |
| P. expansum |
PeAfpA |
A. alternata, Aspergillus spp., Byssochlamys spp., Fusarium spp.,
Penicillium spp. |
[41] |
| P. expansum |
PeAfpB |
Alternaria spp., Aspergillus spp., Byssochlamys spp., Fusarium spp.,
Penicillium spp. |
[41] |
| P. expansum |
PeAfpC |
A. flavus, Byssochlamys spp. |
[41] |
Remarkably, the efficacy of some AFPs in in vivo experiments has been proven. For instance, PgAFP efficiently reduced counts of
A. flavus inoculated on a dry-fermented sausage
[40], while
A. giganteus AFP protected tomato seedlings from vascular wilt disease caused by
F. oxysporum f. sp.
lycopersici [58]. Also PdAfpB and PeAfpA controlled the growth of
P. expansum in apple fruits
[71].
3.2. Antifungal AMPs from Plants
Plant AMPs are constitutively expressed in both plant storage and reproductive organs, but they can also be locally or systematically induced during plant defense response
[72]. Antifungal AMPs have been isolated from a wide variety of plant species, and classified by amino acid sequence, position and number of cysteine residues involved in the disulfide bridges, and/or function to families
[3][73]. A large list of these families show inhibitory activity against mycotoxin-producing fungi (
Table 2). It is noteworthy that, contrary to that described for fungal AFPs, most fungi sensitive to plant antifungal AMPs are
Fusarium species, especially
F. culmurum,
F. graminearum, F. oxysporum and
F. solani. However, other toxigenic fungal species from
Aspergillus (
A. flavus, A. niger),
Penicillium (
P. expansum) and
Alternaria (
A. alternata, A. solani) have been successfully inhibited by antifungal AMPs from plants
[3][74][73]. As a practical example, we highlight the application of the onion (
Allium cepa) defensin
Ace-AMP1on tomato leaves. Treated leaves showed enhance resistance to the tomato pathogen
A. solani (TeA and AOH producer), making this AMP a promising fungicide to be used in agriculture
[75] (
Table 2).
Table 2. Plant antifungal proteins and peptides with activity against mycotoxin-producing fungi.
| Peptide |
Origin |
Target Fungi |
Ref. |
| Defensins |
|
|
|
| Ace-AMP1 |
Allium cepa |
F. solani, F. oxysporum |
[75] |
| Dm-AMP1 |
Dahlia merkii |
Fusarium spp. |
[76] |
| MsDef1 |
Medicago sativa |
F. graminearum |
[77] |
| MtDef4 |
M. truncatula |
F. graminearum |
[78] |
| NaD1, NaD2 |
Nicotiana alata |
F. graminearum, F. oxysporum |
[79] |
| OefDef1.1 |
Olea europea |
Fusarium spp. |
[80] |
| PvD1 |
Phaseolus vulgaris |
F. solani, F. oxysporum |
[81] |
| Rs-AFP2 |
Raphanus sativus |
A. flavus, F. solani |
[82] |
| TPP3 |
N. tabacum |
Fusarium spp. |
[82] |
| Hevein-type |
|
|
|
| Ee-CBP |
Euonymus europaeus |
F. culmorum |
[83] |
| GAFP |
Ginkgo bilolba |
F. graminearum |
[84] |
| SmAMP3 |
Stellariamedia |
F. solani |
[85] |
| Vaccatides |
Vaccaria hispanica |
Fusarium spp. |
[86] |
| WAMP-1a and b |
Triticum aestivum |
F. moniliforme |
[87] |
| Napin |
|
|
|
| BoNap |
Brassica oleracea |
F. culmorum, P. expansum |
[88] |
| Snakins |
|
|
|
| Snakin Z |
Jujube fruits |
A. niger |
[89] |
| SN1, SN2 |
Solanum tuberosum |
F. solani, F. culmorum |
[90] |
| StSN1-2 |
S. tuberosum |
Fusarium spp., A. flavus |
[91] |
| Thaumatin-like |
|
|
|
| Osmotin |
N. tabacum |
F. solani, F. oxysporum |
[57] |
| Zeamatin |
Zea mays |
F. solani |
[92] |
| Thionins |
|
|
|
| Pth-St1 |
S. tuberosum |
F. solani |
[93] |
| Thionin 2.4 |
Arabidopsis thaliana |
F. graminearum |
[94] |
| Tu-AMP1, AMP2 |
Tulipa gesneriana |
F. oxysporum |
[95] |
| Viscotoxin A3 |
Viscum album |
F. solani |
[96] |
| 2S albumin |
|
|
|
| Bn-2S |
Brassica napus |
F. culmorum, F. oxysporum |
[97] |
| CW-1 |
Malva parviflora |
F. graminearum |
[98] |
| Pe AFP1 |
Passiflora edulis |
F. oxysporum |
[99] |
| Pf2 |
P. edulis |
F. oxysporum |
[100] |
| LTPs |
|
|
|
| Bc-nsLTP |
B. campestris |
F. oxysporum |
[101] |
| Ca-LTp1 |
Capsicum annuum |
F. oxysporum |
[102] |
| Ha-AP10 |
Helianthus annus |
F. solani |
[103] |
| Knottins |
|
|
|
| Mj AMP2 |
Mirabilis jalapa |
F. oxysporum |
[104] |
| PAFP-s |
Phytolacca american |
F. oxysporum, F. graminearum |
|
| Hairpinins |
|
|
|
| Sm-AMP-x2 |
Stellaria media |
F.oxysporum, A. niger, A. alternata |
[105] |
| Puroindolines |
|
|
|
| PIN-A |
T. aestivum |
F. culmorum |
[106] |
| PIN-B |
Hordeum vulgare |
F. graminearum |
|
| Gly-rich peptides |
|
|
|
| Gc-GRP |
Coffea canephora |
F. oxysporum |
[107] |
| Pg-AMP1 |
P. edulis |
F. oxysporum |
[108] |
3.3. Antifungal AMPs from Animal Origin
Animal antifungal AMPs are produced at the sites that are constantly exposed to microbes, such as skin and mucosal barriers
[109]. Various antifungal AMPs have been isolated from invertebrates and vertebrate species, including fish, amphibians, and mammals (
Table 3). Several invertebrate AMPs display activity against mycotoxin-producing fungi, in particular
Aspergillus and
Fusarium species, and have been isolated from organisms such as scorpions, silk moth, fruit fly, mantis, bee, termites and ticks. Recently, the susceptibility of the AOH producer
Alternaria brassicicola to thanatin, produced by the spined soldier bug
Podisus maculiventris, was described
[110]. An example of antifungal AMP from fish is pleurocidin, a cationic peptide isolated from the winter flounder
Pleuronectes americanus, which showed antifungal activity against
F. culmorum (DON, NIV and ZEA producer) and
A. niger (OTA producer)
[111]. Finally, mammalian antifungal AMPs are found in human and bovine, and show activity against a large list of mycotoxin-producing fungi including
F. culmurum (DON, NIV, T-2 and ZON producer),
P. expansum (PAT and CIT producer),
A. niger (OTA producer),
A. nidulans (ST),
F. oxysporum (T-2 toxin, HT-2 toxin producer) and
A. flavus (AFs producer).
Of note is the antifungal activity of the human β-defensin 3 (HBD-3) in cereal-based products. Application of 80 μg/mL delayed growth of
F. culmorum,
P. expansum and
A. niger on bread after more than 13 days
[112]. Antifungal functions of bovin lactoferrin and derived peptides have been also reported
[113][114]. Different mycotoxin-producing fungi from
Alternaria, Aspergillus,
Penicillium and
Fusarium were sensitive to lactoferrin-derived peptides. This report is interesting because lactoferrin has been designated by the United States Food and Drug Administration (FDA) as a GRAS food additive
[115].
Table 3. Animal antifungal proteins and peptides with activity against mycotoxin-producing fungi.
| Origin |
Peptide |
Target Fungi |
Ref. |
| Invertebrate |
|
|
|
| Acanthoscurria gomesiana |
Gomesin |
Fusarium spp. |
[116] |
| Bombyx mori |
Cecropin A |
Aspergillus spp., Fusarium spp. |
[117] |
| Centruroides sculpturatus |
BmKbpp2 |
F. culmorum |
[118] |
| Drosophila melanogaster |
Drosomycin |
Fusarium spp., Aspergillus spp. |
[119] |
| D. melanogaster |
Metchnikowin |
F. graminearum |
[120] |
| Heliothis virescens |
Heliomicin |
Fusarium spp. |
[121] |
| Ixodes ricinus |
DefMT3, DefMT5, DefMT6 |
F. graminearum, F. culmorum |
[122] |
| Opistophtalmus carinatus |
Opistoporin-1 |
F. culmorum |
[123] |
| Penaeid shrimps |
Penaeidins |
Aspergillus spp., F. oxysporum |
[124] |
| Podisus maculiventris |
Thanatin |
A. brassicicola, F. culmorum |
[110] |
| Pseudacanthotermes spiniger |
Termicin/Spinigerin |
Aspergillus spp., F. culmorum, F. oxysporym |
[125] |
| Sphodromantis viridis |
Mastoparan-S |
F. culmorum, A. niger, A. fumigatus |
[126] |
| Fish and Amphibians |
|
|
|
| Phyllomedusa bicolor |
Skin-PYY |
A. niger |
[127] |
| Pleuronectes americanus |
Pleurocidin |
F. oxysporum, A. niger, Alternaria spp. |
[111] |
| Mammals |
|
|
|
| Bovine |
Cathelicidin BMAP-28 |
Aspergillus spp., Penicillium spp. |
[128] |
| Bovine |
Indolicidin |
A. niger, Penicillium spp. |
[129] |
| Bovine |
Lactoferrin |
A. niger |
[113] |
| Human |
Defensin HBD-3 |
F. culmorum, P. expansum, A. niger. |
[112] |
| Human |
Hepc20/Hepc25 |
A. niger |
[130] |
| Human |
Tritrptcin |
A. flavus |
[131] |
3.4. Synthetic Antifungal Peptides
The development of synthetic peptides has grown to overcome some drawbacks associated with natural peptides, including low antifungal activity, toxicity or instability. Synthetic AMPs that show antifungal properties against mycotoxin producers are listed in Table 4. This includes analogs of natural AMPs and de novo peptides together with information about the susceptible mycotoxin-producing fungi to these synthetic AMPs, which include different species from Aspergillus, Penicillium and Fusarium genera.
Different strategies have been employed for developing analogs of AMPs. Natural proteins and peptides can be used for the design of novel synthetic bioactive peptides that are more potent than the original ones. They can derive from natural cleavage of natural proteins such as LfcinB17-31 and LfcinB20-25, which are derived from bovine lactoferrin
[114]. Another strategy is to use the sequence of natural occurring AMPs as a template and design a new molecule. For instance, it has been shown that one of the functional regions of defensins is primarily located in the C-terminal β-sheet domain, called the γ-core motif. This is the case of the γ-core motif of the tick selected defensins (DefMT3, DefMT6, and DefMT7), which enhanced antifungal activity against
F. graminearum and
F. culmorum [122][132]. Another extensively used method is based on designing peptides that changes positive charge. In the peptide AGM182 the second disulfide linkage of tachyplesin1 has been replaced by a sequence that assumes an amphipathic β-sheet conformation with maximized positive charge density
[133].
In addition to these analogs of AMPs, many synthetic peptides have been constructed via de novo synthesis such as a group of peptides, named PAFs, which have been designed using a combinatorial library
[134][135][136]. Although these peptides were identified through a nonbiased approach, they show properties of natural AMPs. In fact, PAF26 has been proposed as a model peptide for the characterization and study of cationic, cell-penetrating antifungal peptides
[137].
A good practical example of application is the tachyplesin1-derived peptide AGM182, which caused up to 72% reduction in
A. flavus growth/infection after its expression in transgenic maize plants. Furthermore, reduced fungal growth in the AGM182 transgenic seeds resulted in a significant reduction in AF levels (76–98%)
[138].
Table 4. Synthetic antifungal peptides with activity against mycotoxin-producing fungi.
| Peptide |
Source |
Target Fungi |
Ref. |
| AGM182 |
Tachyplesin-derived |
A. flavus |
[133] |
| Di-K19Hc |
Halocidin-derived |
F. oxysporum, A. niger |
[139] |
| D4E1 |
Cecropin-derived |
Aspergillus spp., Fusarium spp. |
[117][140] |
| γ-core |
DefMT3, DefMT6, DefMT7-derived |
F. graminearum, F. culmorum |
[122][132] |
| K18M |
Thanatin (8–21)-derived |
F. culmorum |
[141] |
| LfcinB17-31/LfcinB20-25 |
Lactoferricin-derived |
A. nidulans, F. oxysporum, P. expansum, Alternaria spp. |
[136] |
| MsrA1 |
Cecropin: Melittin -derived |
F. solani |
[142] |
| BP22 |
de novo |
P. expansum |
[143] |
| D-V13K |
de novo |
Aspergillus spp. |
[144] |
| (KW)n/(RW)n |
de novo |
F. solani, F. oxysporum |
[145] |
| O3TR/C12O3TR |
de novo |
F. culmorum, P. expansum, A. niger |
[146] |
| PAF26/PAF32 |
de novo |
Penicillium spp., F. oxysporum, |
[33][136] |
| PAF76/PAF77 |
de novo |
F. oxysporum |
[147] |
| PEP 6 |
de novo |
F. oxysporum |
[147] |
| PPD1/66-10/77-3 |
de novo |
A. flavus, A. parasiticus |
[148] |