Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mohammad Basyuni | + 2880 word(s) | 2880 | 2022-01-11 02:56:55 | | | |
| 2 | Peter Tang | Meta information modification | 2880 | 2022-01-12 07:34:39 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Basyuni, M. Types of COVID-19 Vaccines. Encyclopedia. Available online: https://encyclopedia.pub/entry/18060 (accessed on 17 January 2026).
Basyuni M. Types of COVID-19 Vaccines. Encyclopedia. Available at: https://encyclopedia.pub/entry/18060. Accessed January 17, 2026.
Basyuni, Mohammad. "Types of COVID-19 Vaccines" Encyclopedia, https://encyclopedia.pub/entry/18060 (accessed January 17, 2026).
Basyuni, M. (2022, January 11). Types of COVID-19 Vaccines. In Encyclopedia. https://encyclopedia.pub/entry/18060
Basyuni, Mohammad. "Types of COVID-19 Vaccines." Encyclopedia. Web. 11 January, 2022.
Copy Citation
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a highly contagious virus that emerged at the end of 2019 and has caused an upper respiratory disease pandemic, currently known as Coronavirus Disease 2019 (COVID-19). Vaccine clinical studies are developing promptly with the aim of obtaining vaccines that are effective in suppressing the spread of the virus; however, the development of viral mutations raises concerns about the decreasing effectiveness of the resulting vaccine, which also results in the need for more in-depth studies. There have been 330 vaccines developed, including 136 clinical developments and 194 pre-clinical developments.
COVID-19
mutated SARS-CoV-2 virus
vaccine
post vaccine surveillance
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a highly contagious virus that emerged at the end of 2019 and has caused an upper respiratory disease pandemic, currently known as Coronavirus Disease 2019 (COVID-19) [1]. On 11 March 2020, WHO declared COVID-19 a global pandemic, and it was reported that COVID-19 had spread in 197 countries (on 25 March 2020) with a mortality rate of 18,440 and a recovery rate of 114,802 globally [2]. Efforts to defeat the COVID-19 pandemic continue to date, and vaccinations have been implemented globally.
Until now, the SARS-CoV-2 virus continues to mutate, and there are several variants that have become a variant of concern (VOC), including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta). At the end of 2020, new variants emerged, namely C.37 (Lambda) and B.1.621 (Mu), which were included in the Variants of Interest (VOI) category [3]. The Alpha variant is estimated to have a 61% (42–82%) higher risk of death than the pre-existing variants, with greater infectiousness and disease severity [4]. P.1, another highly contagious variant, has been circulating in Brazil since the middle of 2020. This variant has been linked to an outbreak of infections in Manaus, Brazil’s Amazon, which has put the healthcare system on the verge of collapsing. B.1.351 was discovered late last year in South Africa [5], while the Delta variant is known to be notoriously contagious. Within five weeks of its discovery in April–June 2021, the Delta variant became the dominant SARS-CoV-2 variant in Mesa County, Colorado, and is now the dominant variant in the United States [6].
2. SARS-CoV-2 and Receptors
SARS-CoV-2 is a subfamily of Coronaviridae (CoV), part of the Coronaviridae RNA virus family. The two types of CoV viruses that have caused serious illness are SARS-CoV-1 that caused SARS in 2002–2003, and MERS-CoV causing MERS that occurred in the Middle East in 2012 [7]. SARS-CoV-2 was first identified by independent Chinese scientists through the bronchoalveolar lavage fluid of a patient who had severe pneumonia at the start of a COVID-19 case in Wuhan [1]. The identification results presented the presence of beta-coronavirus that showed 85% genomic similarity with the Bat-SARS-like CoV virus (bat-SL-CoVZC45, GenBank: MG772933.1); afterward, the virus was isolated and named 2019-nCoV. The electron micrograph from 2019-CoV shows the sphere shape with various diameters between 60–140 nm, with a clear spike between 9–12 nm in size; thus, it looks like a solar corona [8].
The SARS-CoV-2 spike protein is 1273 amino acids long, a little longer than SARS-CoV (1255 amino acids) and Bat-SARS-CoV (1245–1269 amino acids). The China National Center for Bioinformation’s 2019 Novel Coronavirus Resource identified a variant of the SARS-CoV-2 strain, of which 15,018 mutations were discovered worldwide [1]. Visualization of SARS-CoV-2 with transmission electron microscopy exhibited the transcriptomics of SARS-CoV-2, presented in Figure 1A–C [9].
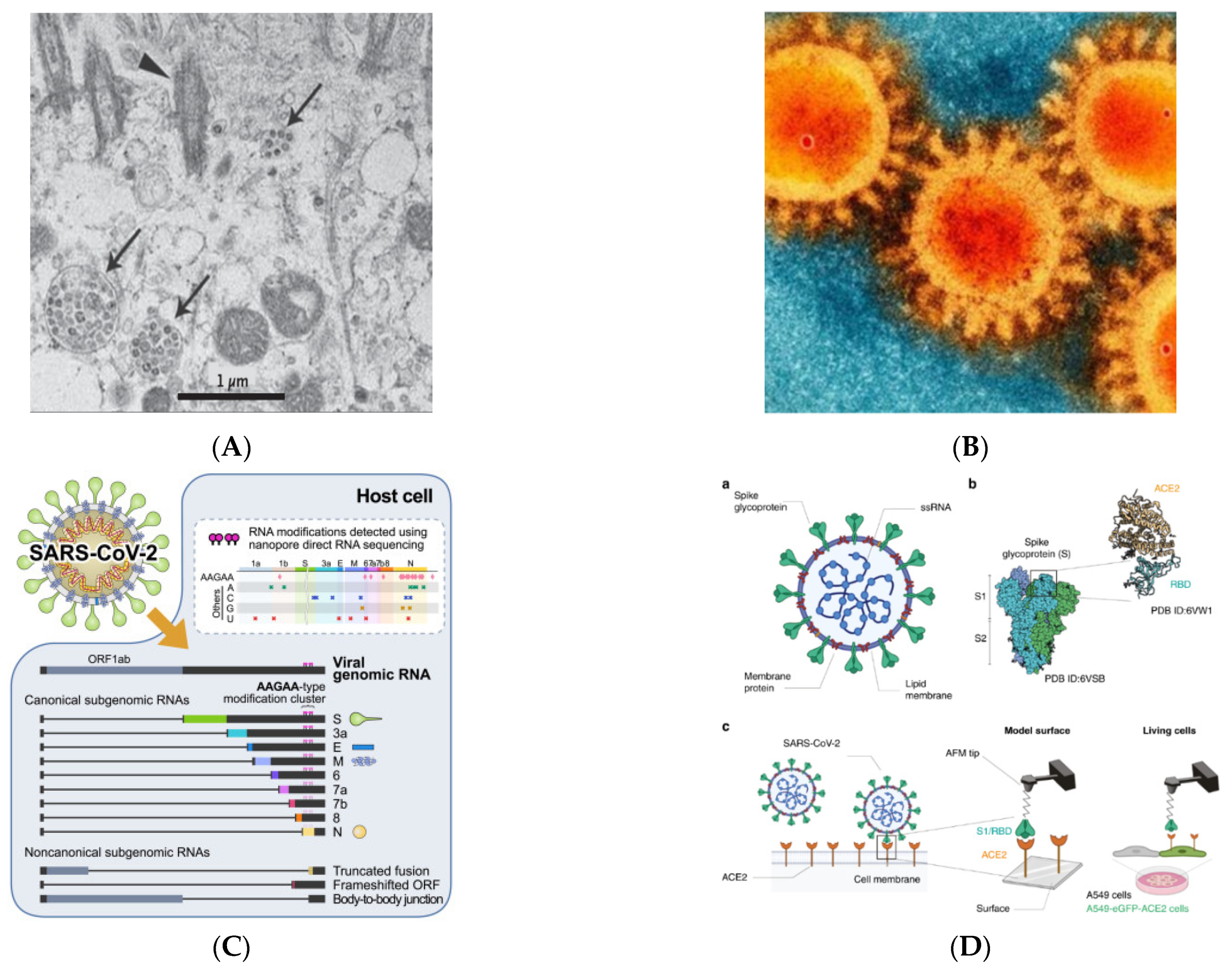
Figure 1. Visualization of SARS-CoV-2 (adapted with permission from Zhu et al., 2019; Bharadwaj et al., 2020; Kim et al., 2020 and Yang et al., 2020) [8][9][10][11] (A) Visualization of SARS-CoV-2 with Transmission Electron Microscopy (Virus particles in the human airway epithelial cell). Arrowheads represent extracellular virus particles, while arrows represent inclusion bodies generated by virus components; (B) SEM image of SARS-CoV-2 (Large protrusions emerged out from the spike of viral surface, which forms a crown-like appearance, that’s why given the name ‘coronavirus’ (Latin word means crown)); (C) Transcriptomic (The full-length genomic RNA and nine major subgenomic RNAs); (D) SARS-CoV-2—ACE2 interaction (The initial attachment of SARS-CoV-2 to cells involves specific binding between the viral S glycoprotein and the cellular receptor, ACE2. The interactions are monitored by AFM on model surfaces, where the ACE2 receptor is attached to a surface and the S1 subunit of the RBD onto the AFM tip, and on A549 living cells expressing or not fluorescently labeled ACE2). a. A SARS-CoV-2 particle, an enveloped ssRNA virus with the spike glycoprotein (S) on its surface that mediates binding to host cells, is depicted schematically. b. A complex between the receptor-binding domain (RBD, a subunit of the S glycoprotein) and the ACE2 receptor has previously been discovered by structural research. c. A diagram depicting the use of AFM to investigate SARS-CoV-2 binding.
SARS-CoV-2 shares a 79 percent genome sequence similarity with SARS-CoV, and a 50 percent similarity with MERS-CoV. Its genome organization is similar to that of other beta-coronaviruses. The six functional open reading frames (ORFs) are arranged from 5′ to 3′: replicase (ORF1a/ORF1b), spike (S), envelope (E), membrane (M), and nucleocapsid (N). The majority of the proteins encoded by SARS-CoV-2 are similar in length to the corresponding proteins in SARS-CoV. Except for the S gene, which differs, SARS-CoV-2 shares more than 90% amino acid identity with SARS-CoV [1]. The replicase S gene encodes a huge polyprotein (pp1ab) that is proteolytically degraded into 16 non-structural proteins involved in transcription and viral replication. The majority of these non-structural SARS-CoV-2 proteins share more than 85% of their amino acid sequence with SARS- CoV-2 [1].
SARS-CoV-2, like SARS-CoV, binds to the human angiotensin-converting enzyme 2 (ACE2) receptor. ACE-2 is a membrane receptor protein that is primarily found in adipose tissue, the kidney, the heart, and the small intestine. In humans, SARS-CoV-2 infection causes a range of symptoms from mild to severe respiratory failure. When SARS-CoV-2 binds to airway epithelial cells, it replicates, migrates, and enters alveolar epithelial cells in the lungs. This rapid replication causes a strong immune response known as a cytokine storm, which results in acute respiratory distress syndrome and respiratory failure, which are the leading causes of death in COVID-19 patients [12][13][14]. The upper respiratory tract provides the first line of defense in that it activates a very complex system of signaling and recruitment of immune cells in order to prevent pathogens from reaching the alveoli, the most important part of the respiratory system, as they are responsible for gas exchange. Nevertheless, SARS-CoV-2 or other coronavirus are able to reduce or delay the expression of cytokines in human lung epithelial cell lines. This is an effective system to escape immune recognition by innate receptors in the infected cell, which could therefore facilitate the progression of the virus into alveolar epithelial cells [15].
Previous research has used the force-distance (FD) curve-based atomic force microscopy to investigate the biophysical properties of the SARS-CoV-2 S-glycoprotein binding to ACE2 receptors on model surfaces and in living cells (FD-curve-based AFM). They extracted the kinetics and thermodynamics of the in vitro interactions and compared the binding properties of both the S1 subunit and the RBD. Both have been tested as potent binding inhibitor peptides targeting the viral S glycoprotein, and a significant reduction in binding properties has been observed. Figure 1D depicts the SARS-CoV-2 scheme as well as the binding scheme with the ACE2 receptor [11]. Coronavirus entrance necessitates the virus’s attachment to the ACE2 receptor on the host cell’s surface, followed by priming by TMPRSS2. The possible SARS-CoV-2 cofactors ACE2, TMPRSS2, and FURIN are expressed largely in bronchial cells switching from secretory to ciliated identity, according to this study, which used previously unreported single-cell data from the human lung and bronchia [16].
3. Mutation in the SARS-CoV-2
Several variants of SARS-CoV-2 are circulating globally and have been identified, including the B.1.1.7 (Alpha) from the United Kingdom, B.1.351 (Beta) from South Africa, B.1.617.2 (Delta) from India, P.1 (Gamma) from Brazil, and B.1.1.529 (Omicron). The Alpha variant came from the UK and was first identified in December 2020 [17][18]. Although distinct, the Alpha and Beta variants share common characteristics, including known escape mutations discovered in vitro through antibody pressure selection [19]. The SARS-CoV-2 B.1.617.2 Variant of Concern (VOC) or Delta variant, first detected in India, has now displaced the B.1.1.7 (Alpha) strain, which emerged in the UK with the second COVID-19 wave in late 2020. The B.1.617.2 variant may be transmitted at a higher rate than other variants [17][20]. The WHO Technical Advisory Group on SARS-CoV-2 Virus Evolution recognized the B.1.1.529 COVID-19 variant, which was first detected in Botswana and South Africa, as the Omicron variant of concern on 26 November 2021. Omicron, the SARS-CoV-2 variant responsible for a cluster of cases in South Africa and that is now spreading around the world, is the most heavily mutated variant to emerge so far and carries mutations similar to changes seen in previous variants of concern associated with enhanced transmissibility and partial resistance to vaccine-induced immunity [21].
The WHO and the NIAID directors conveyed to the public and health practitioners about a new variant called “Mu” in the COVID Briefing on 2 September 2021. The Mu-variant is a SARS-CoV-2 variant named B.1.621, according to PANGO lineage, which was first identified in a sample of 11 January 2021 in Colombia [22]. The status of Mu-variant in the world is designated as a VOI by WHO on 30 August 2021. According to WHO data, the distribution of Mu-variant in the world as of 1 September 2021 has been found in at least 39 countries [3]. The Mu-variant carried at least 21 point mutations in the genetic material of SARS-CoV-2, of which nine were in the viral spike protein. This variant carries several key mutations previously known in other variants. The key mutation carried by the Mu-variant is N501Y (as in the Alpha-variant); E484K (as in the Beta-variant); P681H (as in the Delta-variant) [23].
WHO decided to monitor this variant because the combination of mutations carried by the Mu-variant has the potential to decrease antibody neutralization, as shown in early studies of convalescent plasma and vaccine sera (however, the scientists from the Virus Evolution Working Group agree that further research is needed). Its global prevalence continues to decline (currently < 0.1%); however, this variant is still under scrutiny due to its consistent development in Ecuador and Colombia [3][24]. The WHO Virus Evolution Working Group (now called the Technical Advisory Group on Virus Evolution) released recent enhancing readiness for Omicron (B.1.1.529): technical brief and priority actions for WHO members on 10 December 2021. It has been mentioned that the current understanding of the Omicron variant based on recent data is likely to evolve as more data becomes available.
4. Types of COVID-19 Vaccines
Over the last few decades, advances in molecular biology and vaccinology have resulted in the development of a diverse range of novel vaccine platform technologies. These platform technologies include live pathogen inactivation and targeted attenuation to the delivery of synthetic peptide antigens and recombinant protein antigens, as well as virus-like particles (VLPs), non-replicating and replicating viral vectors, polysaccharide-protein conjugates, and nucleic acid-based (DNA and RNA) vaccines. Many of the currently marketed vaccines against infectious diseases are based on these platform technologies [25][26]. The COVID-19 Vaccine Development Platform consists of the following components: inactivated whole SARS-CoV-2, DNA-based vaccine (plasmid DNA expressing S protein), m-RNA-based vaccine (receptor binding domain of spike protein), subunit vaccine (recombinant spike protein), and vector-based vaccine (replicating or non-replicating viral vector used for the delivery of spike protein). Figure 2 depicts the COVID-19 Vaccine Development Platform [27].
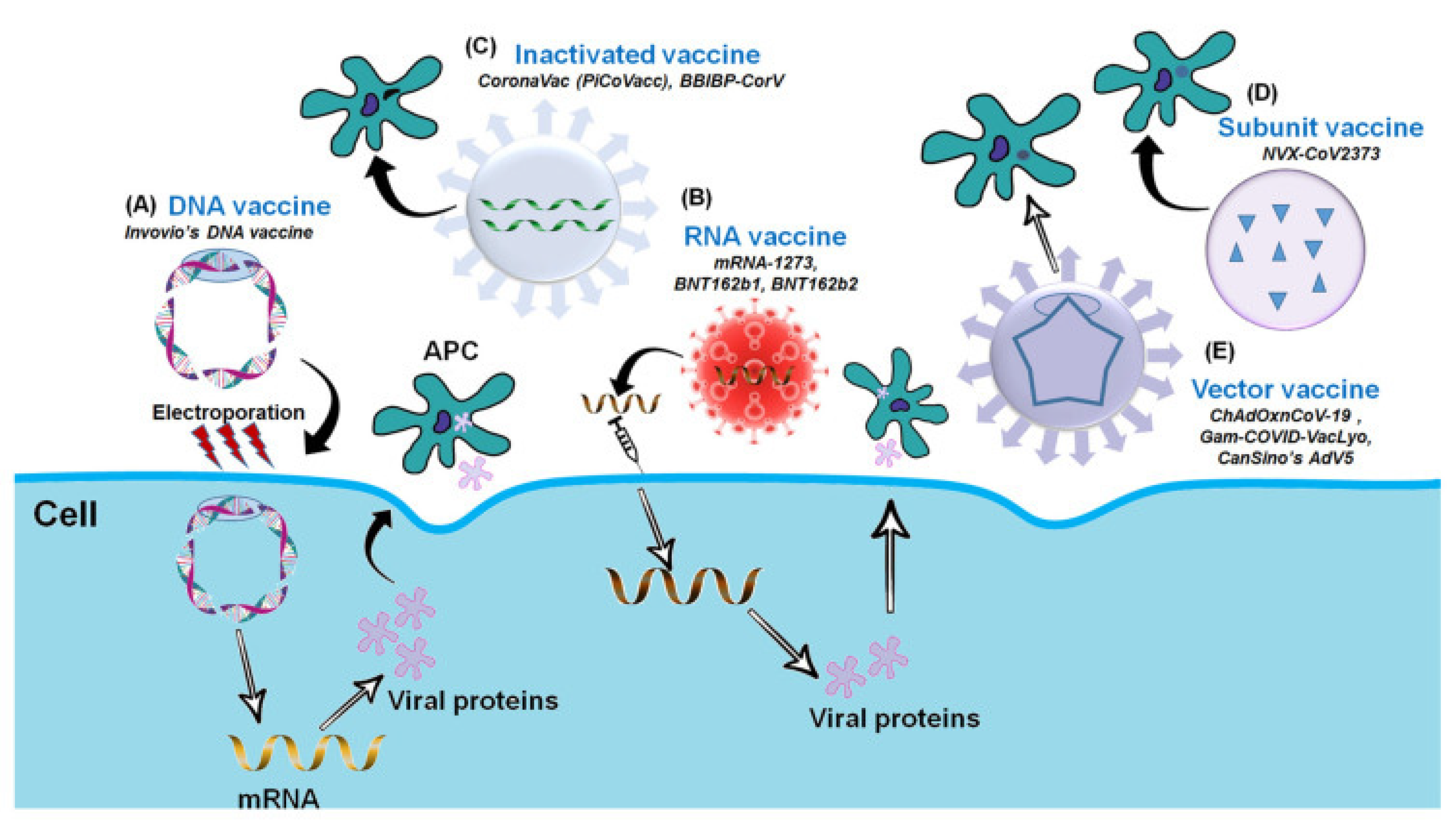
Figure 2. The Platform for COVID-19 Vaccine (adapted with permission from Ashraf et al., 2021) [27]. Platforms for the COVID-19 Vaccine Development. (A) DNA vaccine: Plasmid DNA expressing S protein. (B) RNA vaccine: mRNA-based (RBD of S-protein). (C) Inactivated vaccine: Inactivated whole SARS-CoV-2. (D) Subunit vaccine: Recombinant S-protein and (E) Vector-based vaccine: Replicating or Non-replicating viral vector used for the delivery and expression of S protein.
Based on data from WHO as of 7 December 2021, there are currently 330 COVID-19 vaccines under development, with 136 vaccine candidates having entered the clinical-phase trial and 194 others still in the pre-clinical phase. Among the vaccine candidate platforms currently being tested are protein subunit 47 (35%), viral vector non-replicating 20 (15%) and replicating 2 (1%), DNA based vaccines 15 (11%), mRNA based vaccines 22 (16%), and inactivated virus 13 (13%). According to the 136 types of vaccines that have been clinically tested, 10 vaccines have reached stage 4 clinical trials [28].
4.1. Inactivated Virus-Based Vaccines
Pre-clinical studies have been conducted on a number of COVID-19 vaccine candidates based on well-established technology. This method is based on an existing technology platform for pathogen inactivation in blood products, in which ultraviolet light and riboflavin are used to inactivate the virus through targeted damage to nucleic acids, while proteins and viral antigens are preserved. This technique has been shown to be effective in inactivating MERS-CoV. The development of conventional inactivated vaccines necessitates the cultivation of high titers of infectious virus, which, in the case of SARS-CoV-2, must take place in biosafety level 3 facilities, posing a major safety concern. Furthermore, incomplete virus inactivation poses a potential risk to vaccine production workers, as well as the possibility of disease outbreaks in vaccinated populations and the induction of harmful immune or inflammatory responses [26][29].
CoronaVac is a vaccine with an inactivated virus-based platform. This vaccine has varying efficacy, with the latest efficacy results from phase 3 clinical trials in Indonesia showing this vaccine is quite effective with 65.3% efficacy after the second dose of 1620 participants aged 18–59 years [30].
4.2. DNA-Based Vaccines
The introduction of the DNA vaccine, which encodes for the antigen and an adjuvant that induces the adaptive immune response, has been the most revolutionary approach to vaccination. The transfected cells express the transgene, supplying a steady supply of transgene-specific proteins that are very similar to the live virus [31]. In the form of DNA, this vaccination comprises a subset of the virus’s genes. The DNA is employed as a template for in situ expressions of possibly innocuous viral proteins, which trigger a protective immune response after injection. The safety and scalability for large production are two of the most significant advantages of this type of vaccine.
The DNA has to enter the nucleus in order to be transcribed. The worry arises in terms of the potential long-term risk of tumorigenicity, especially when injected into young people. These concerns are supported by a document issued by the FDA under the title “Long-Term Follow-up after Administration of Human Gene Therapy Products, Guidance for Industry” [32]. In contrast, a 1997 study found that parenteral administration of hybridoma DNA does not result in the development of local tumors. Furthermore, there was no evidence of the hybridoma DNA’s systemic carcinogenic potential [33]. Behind the conflicting DNA-based vaccine platforms, there are several vaccines being developed, including INO-480+electroporation, nCov vaccine, AG0301-COVID19, and GX-19N [28].
4.3. mRNA-Based Vaccines
RNA vaccines contain virus genes in the form of mRNA, which is then translated into viral proteins after cytosolic delivery. The mRNA is a new, non-infectious, and non-integrating platform with almost no risk of insertional mutagenesis. Currently, non-replicating RNA and virus-derived self-replicating RNAs are being studied. The immunogenicity of the mRNA can be reduced, and changes can be made to improve the stability of these vaccines [26][31].
The mRNA-1273 vaccine, developed by Moderna Therapeutics (Cambridge, MA, USA) and the National Institute of Allergy and Infectious Disease (NIAID), was the first candidate vaccine to enter Phase I clinical testing, just 42 days after the full SARS-CoV-2 genome was sequenced (ClinicalTrials.gov identifier NCT04283461) [34][35].
Recent studies of the BNT162b2 vaccine have shown that after 6 months of use, the vaccine is safe and has acceptable adverse events. The efficacy of this vaccine reaches 91.3% and has a high efficacy advantage in dealing with beta variants [36]. In phase 4 clinical trials, the vaccine was tested as a booster or third dose in participants aged over 16 years and patients with multiple sclerosis [37][38][39].
4.4. Protein Subunit-Based Vaccine
Vaccines based on synthetic peptides or recombinant antigenic proteins are known as subunit vaccines. Subunit vaccines have low immunogenicity and require adjuvant support to enhance the vaccine-induced immune response. Subunit vaccines contain only specific viral antigenic fragments and no additional pathogenic virus components. As a result, this method eliminates concerns about incomplete viral inactivation, virulence recovery, and pre-existing anti-vector immunity. As a result, subunit vaccines are widely regarded as extremely safe. Furthermore, subunit vaccines can specifically target well-characterized neutralizing antigenic epitopes and improve immunogenicity and/or efficacy when combined with adjuvants. Adjuvants can extend the antigenic material’s biological half-life or improve the immunomodulatory cytokine response [26].
Vaccines developed with this model include NVX-CoV2373 (Novavax, Inc.). In an animal model, it demonstrated high immunogenicity by measuring anti-spike antibodies, which prevent the spike protein from attaching to the receptor, as well as wild-type virus-neutralizing antibodies [31]. Adult participants were given a two-dose regimen of the NVX-CoV2373 vaccine, which provided 89.7 percent protection against SARS-CoV-2 infection and demonstrated high efficacy against the B.1.1.7 variant [40].
4.5. Viral Vector-Based Vaccine
A vaccine based on viral vectors is a promising anti-pathogen treatment. These vaccines are highly specific in terms of delivering genes to target cells, efficient in terms of gene transduction, and, once again, inducing an immune response. They provide a long-term and high level of antigenic protein expression and thus have a high potential for prophylactic use, as these vaccines activate and prime cytotoxic T cells (CTL), resulting in the elimination of virus-infected cells [31][41]. Viral vectors are used to deliver vaccine antigens to target cells or tissues, and both replicating and non-replicating viral vectors are available. Adenoviruses and poxviruses are examples of viral vectors of replicating and non-replicating forms.
One of the viral vector vaccine platforms that have been used today is ChAcOx1 nCoV-19 (AstraZeneca and Oxford). Based on clinical trial data from 17,177 people consisting of 8948 in the UK, 6753 in Brazil, and 1476 in South Africa, showed high safety and efficacy in participants aged 18–55 years [42].
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154.
- Baloch, S.; Baloch, M.A.; Zheng, T.; Pei, X. The Coronavirus Disease 2019 (COVID-19) Pandemic. Tohoku J. Exp. Med. 2020, 250, 271–278.
- WHO. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants (accessed on 5 September 2021).
- Davies, N.G.; Jarvis, C.I.; CMMID COVID-19 Working Group; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021, 593, 270–274.
- Burki, T. Understanding variants of SARS-CoV-2. Lancet 2021, 397, 462.
- Herlihy, R.; Bamberg, W.; Burakoff, A.; Alden, N.; Severson, R.; Bush, E.; Kawasaki, B.; Berger, B.; Austin, E.; Shea, M.; et al. Rapid Increase in Circulation of the SARS-CoV-2 B.1.617.2 (Delta) Variant—Mesa County, Colorado, April–June 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 1084–1087.
- Platto, S.; Wang, Y.; Zhou, J.; Carafoli, E. History of the COVID-19 pandemic: Origin, explosion, worldwide spreading. Biochem. Biophys. Res. Commun. 2020, 538, 14–23.
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733.
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10.
- Bharadwaj, A.; Wahi, N.; Saxena, A.; Chaudhary, D. Proteome Organization of COVID-19: Illustrating Targets for Vaccine Development. J. Pure Appl. Microbiol. 2020, 14, 831–840.
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541.
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256.
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613.
- Das, G.; Ghosh, S.; Garg, S.; Ghosh, S.; Jana, A.; Samat, R.; Mukherjee, N.; Roy, R.; Ghosh, S. An overview of key potential therapeutic strategies for combat in the COVID-19 battle. RSC Adv. 2020, 10, 28243–28266.
- Carcaterra, M.; Caruso, C. Alveolar Epithelial Cell Type II as Main Target of SARS-CoV-2 Virus and COVID-19 Development via NF-Kb Pathway Deregulation: A Physio-Pathological Theory. Med. Hyphotheses 2021, 146, 110412.
- Lukkasen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Mesiter, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 Receptor ACE2 and TMPRSS2 Are Primarily Expressed in Bronchial Transient. EMBO J. 2020, 39, e105114.
- Anonymous. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html (accessed on 28 October 2021).
- Firestone, M.J.; Lorentz, A.J.; Wang, X.; Como-Sabetti, K.; Vetter, S.; Smith, K.; Holzbauer, S.; Meyer, S.; Ehresmann, K.; Danila, R.; et al. First Identified Cases of SARS-CoV-2 Variant B.1.1.7 in Minnesota—December 2020–January 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 278–279.
- Planas, D.; Bruel, T.; Grzelak, L.; Guivel-Benhassine, F.; Staropoli, I.; Porrot, F.; Planchais, C.; Buchrieser, J.; Rajah, M.M.; Bishop, E.; et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021, 27, 917–924.
- Dougherty, K.; Mannell, M.; Naqvi, O.; Matson, D.; Stone, J. SARS-CoV-2 B.1.617.2 (Delta) Variant COVID-19 Outbreak Associated with a Gymnastics Facility—Oklahoma, April–May 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1004–1007.
- Torjesen, I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ 2021, 375, n2943.
- Anonymous. Lineage B.1.621. Available online: https://cov-lineages.org/lineage.html?lineage=B.1.621 (accessed on 5 September 2021).
- Anonymous. B.1.621 Lineage Report. Available online: https://outbreak.info/situation-reports?pango=B.1.621 (accessed on 5 September 2021).
- Messali, S.; Bertelli, A.; Campisi, G.; Zani, A.; Ciccozzi, M.; Caruso, A.; Caccuri, F. A cluster of the new SARS-CoV-2 B.1.621 lineage in Italy and sensitivity of the viral isolate to the BNT162b2 vaccine. J. Med. Virol. 2021, 93, 6468–6470.
- Lurie, N.; Saville, M.; Hatchett, R.; Halton, J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020, 382, 1969–1973.
- Frederiksen, L.S.F.; Zhang, Y.; Foged, C.; Thakur, A. The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. Front. Immunol. 2020, 11, 1817.
- Ashraf, M.; Kim, Y.; Kumar, S.; Seo, D.; Ashraf, M.; Bae, Y.-S. COVID-19 Vaccines (Revisited) and Oral-Mucosal Vector System as a Potential Vaccine Platform. Vaccines 2021, 9, 171.
- WHO. Draft Landscape and Tracker of COVID-19 Candidate Vaccines. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 16 December 2021).
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an Inactivated Vaccine Candidate for SARS-CoV-2. Science 2020, 369, 77–81.
- Fadlyana, E.; Rusmil, K.; Tarigan, R.; Rahmadi, A.R.; Prodjosoewojo, S.; Sofiatin, Y.; Khrisna, C.V.; Sari, R.M.; Setyaningsih, L.; Surachman, F.; et al. A phase III, observer-blind, randomized, placebo-controlled study of the efficacy, safety, and immunogenicity of SARS-CoV-2 inactivated vaccine in healthy adults aged 18–59 years: An interim analysis in Indonesia. Vaccine 2021, 39, 6520–6528.
- Kaur, S.P.; Gupta, V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020, 288, 198114.
- FDA. Long Term Follow-Up After Administration of Human Gene Therapy Products, Guidance for Industry; FDA.gov: Silver Spring, MD, USA, 2020.
- Dortant, P.; Claassen, I.; Van Kreyl, C.; Van Steenis, G.; Wester, P. Risk Assessment on the Carcinogenic Potential of Hybridoma Cell DNA: Implications for Residual Contaminating Cellular DNA in Biological Products. Biologicals 1997, 25, 381–390.
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438.
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931.
- Thomas, S.J.; Moreira, E.D., Jr.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Polack, F.P.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine through 6 Months. N. Engl. J. Med. 2021, 385, 1761–1773.
- Cowling, B.J.; The University of Hong Kong. Third Dose of mRNA Vaccination to Boost COVID-19 Immunity. Available online: https://clinicaltrials.gov/ct2/show/NCT05057182 (accessed on 16 December 2021).
- Guanaccia, B.J.; Browne, F.; Yale-Griffin Prevention Research Cente. COVID-19 Booster Vaccination in Persons with Multiple Sclerosis. Available online: https://clinicaltrials.gov/ct2/show/record/NCT05081271 (accessed on 16 December 2021).
- Pfizer. Study to Evaluate the Safety and Efficacy of a Booster Dose of BNT162b2 Against COVID-19 in Participants ≥16 Years of Age. Available online: https://clinicaltrials.gov/ct2/show/NCT04955626?term=vaccine&recrs=abdf&cond=COVID-19&phase=012345&sort=nwst&draw=2 (accessed on 16 December 2021).
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183.
- Thanh Le, T.; Andreadakis, Z.; Kumar, A.; Gómez Román, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306.
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111.
More
Information
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Entry Collection:
COVID-19
Revisions:
2 times
(View History)
Update Date:
12 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




